Search Count: 58
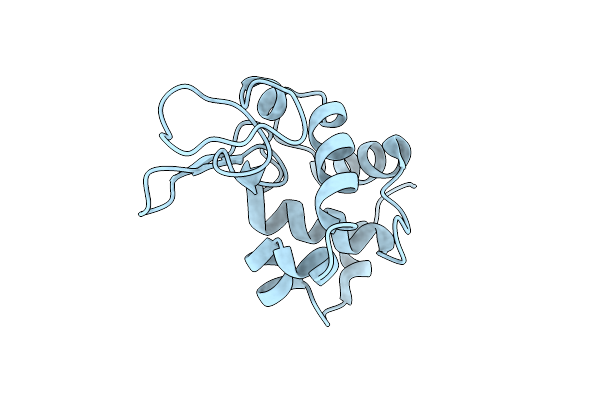 |
Hen Egg-White Lysozyme Structure Determined By 3Ded/Microed On A 200 Kev Microscope
Organism: Gallus gallus
Method: ELECTRON CRYSTALLOGRAPHY Resolution:2.80 Å Release Date: 2025-03-19 Classification: HYDROLASE |
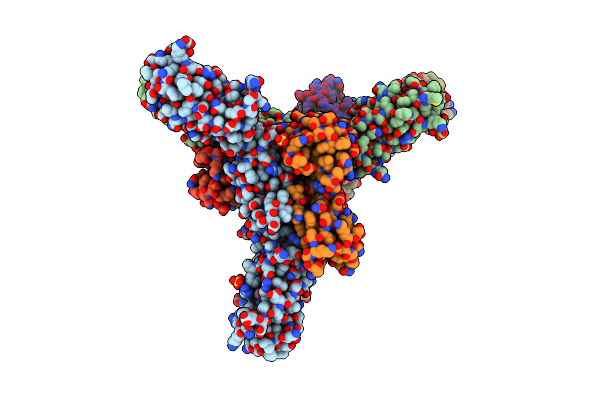 |
Complex Of Diubiquitin-Derived Artificial Binding Protein (Affilin) Variant Af2 With Its Target Oncofetal Fibronectin (Fragment 7B8)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2024-06-26 Classification: DE NOVO PROTEIN Ligands: SO4 |
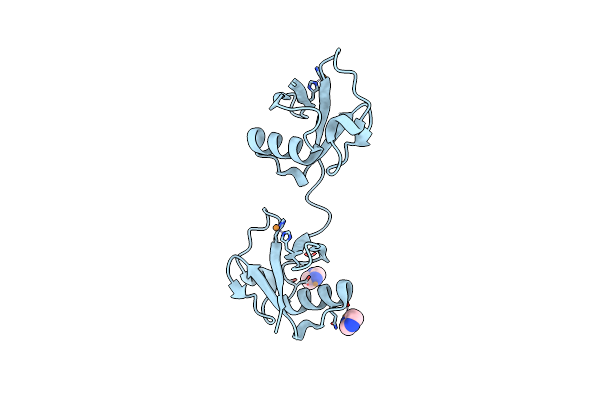 |
Diubiquitin-Derived Artificial Binding Protein (Affilin) Variant Af1 Specific To Oncofetal Fibronectin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-06-26 Classification: DE NOVO PROTEIN Ligands: CU, IMD |
 |
Crystal Structure Of Human Mature Meprin Beta In Complex With The Specific Inhibitor Mwt-S-270
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2021-03-03 Classification: HYDROLASE Ligands: ZN, EDO, RUE, CA, CL, NAG |
 |
Molecular Structure Of Mouse Apoferritin Resolved At 2.7 Angstroms With The Glacios Cryo-Microscope
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Resolution:2.73 Å Release Date: 2020-05-13 Classification: METAL BINDING PROTEIN Ligands: FE, MG |
 |
Organism: Porphyromonas gingivalis w83
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2019-03-06 Classification: TRANSFERASE Ligands: ZN |
 |
Organism: Tannerella forsythia (strain atcc 43037 / jcm 10827 / fdc 338)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-03-06 Classification: TRANSFERASE Ligands: ZN, SO4 |
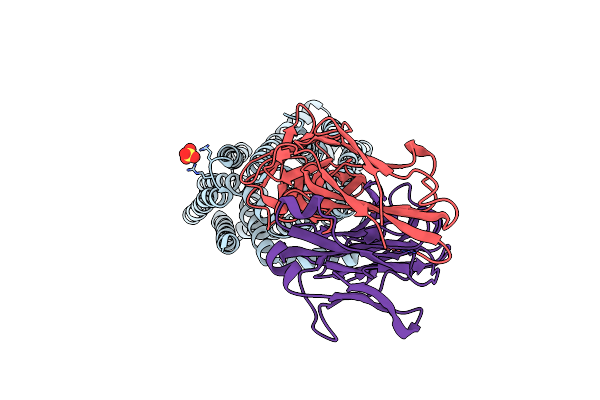 |
Crystal Structure Of E.Coli Multidrug/H+ Antiporter Mdfa In Outward Open Conformation With Bound Fab Fragment
Organism: Escherichia coli k-12, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2018-10-03 Classification: TRANSPORT PROTEIN Ligands: SO4 |
 |
Structure Of Pyroglutamate-Abeta-Specific Fab C#17 In Complex With Human Abeta-Pe3-12Pegb
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2017-06-28 Classification: IMMUNE SYSTEM |
 |
Structure Of Pyroglutamate-Abeta-Specific Fab C#17 In Complex With Murine Abeta-Pe3-18Pegb
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-06-28 Classification: IMMUNE SYSTEM |
 |
Structure Of Pyroglutamate-Abeta-Specific Fab C#6 In Complex With Human Abeta-Pe3-12-Pegb
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2017-06-28 Classification: IMMUNE SYSTEM Ligands: SO4, GOL |
 |
Structure Of Pyroglutamate-Abeta-Specific Fab C#24 In Complex With Human Abeta-Pe3-18
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2017-06-28 Classification: IMMUNE SYSTEM |
 |
Organism: Thymus vulgaris
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-01-13 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-09-02 Classification: SIGNALING PROTEIN Ligands: TRS |
 |
Structure Of The Toll - Spatzle Complex, A Molecular Hub In Drosophila Development And Innate Immunity
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-04-09 Classification: IMMUNE SYSTEM/CYTOKINE Ligands: NAG, EPE |
 |
Structure Of The Toll - Spatzle Complex, A Molecular Hub In Drosophila Development And Innate Immunity (Glycosylated Form)
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2014-04-09 Classification: IMMUNE SYSTEM/CYTOKINE Ligands: NAG, EPE |
 |
Crystal Structure Of Trypsiligase (K60E/N143H/Y151H/D189K Trypsin) Trigonal Form
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2014-02-19 Classification: HYDROLASE Ligands: CA, GOL |
 |
Crystal Structure Of Trypsiligase (K60E/N143H/Y151H/D189K Trypsin) Orthorhombic Form
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2014-02-19 Classification: HYDROLASE Ligands: CA, GOL |
 |
Crystal Structure Of Trypsiligase (K60E/N143H/Y151H/D189K Trypsin) Orthorhombic Form, Zinc-Bound
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2014-02-19 Classification: HYDROLASE Ligands: CA, GOL, ZN |
 |
Crystal Structure Of Trypsiligase (K60E/N143H/Y151H/D189K Trypsin) Complexed To Yrh-Ecotin (M84Y/M85R/A86H Ecotin)
Organism: Bos taurus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2014-02-19 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA, ZN |

