Search Count: 20
 |
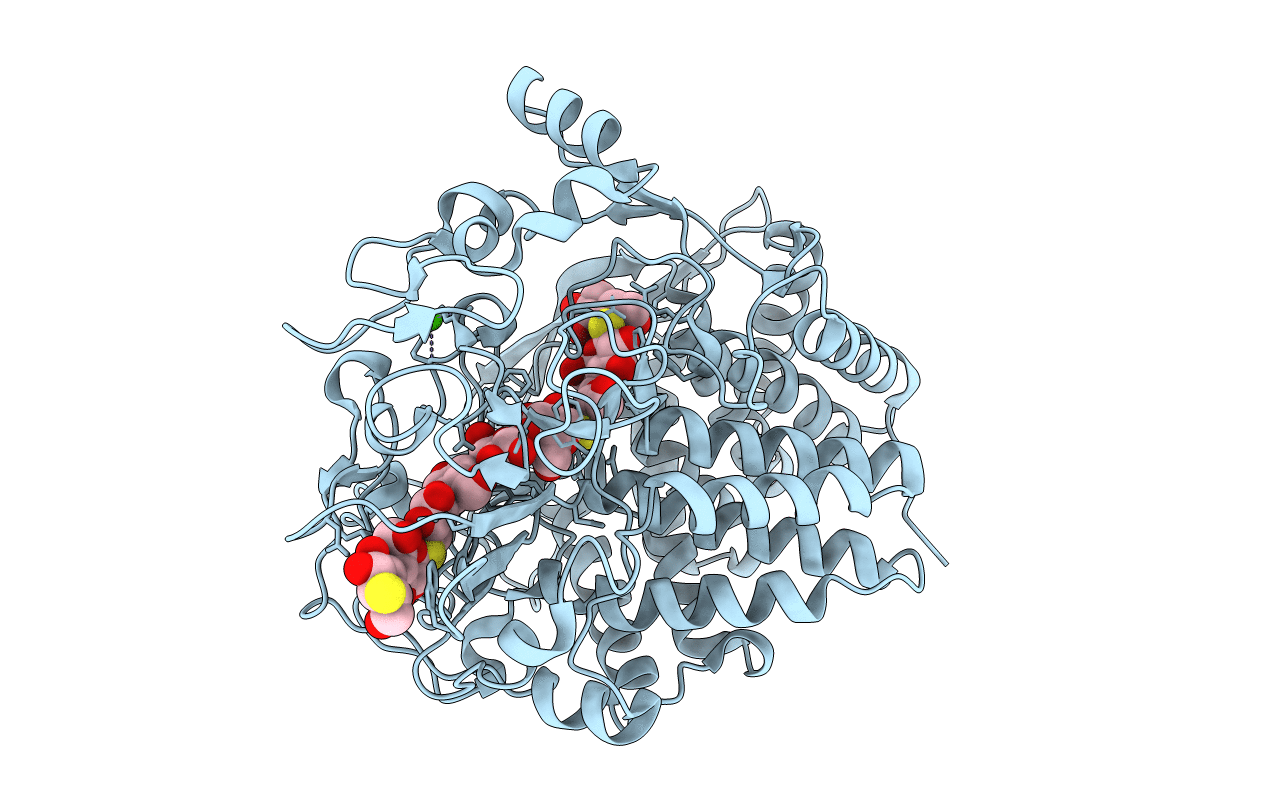
Structure Of Recombinant Human Dnase I (Rhdnasei) In Complex With Magnesium And Phosphate.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-01-09 Classification: HYDROLASE Ligands: CA, MG, PO4, BTB, NAG |
 |
Crystal Structure Of The Mutant E55Q Of The Cellulase Cel48F In Complex With A Thio-Oligosaccharide
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-01-15 Classification: HYDROLASE Ligands: CA |
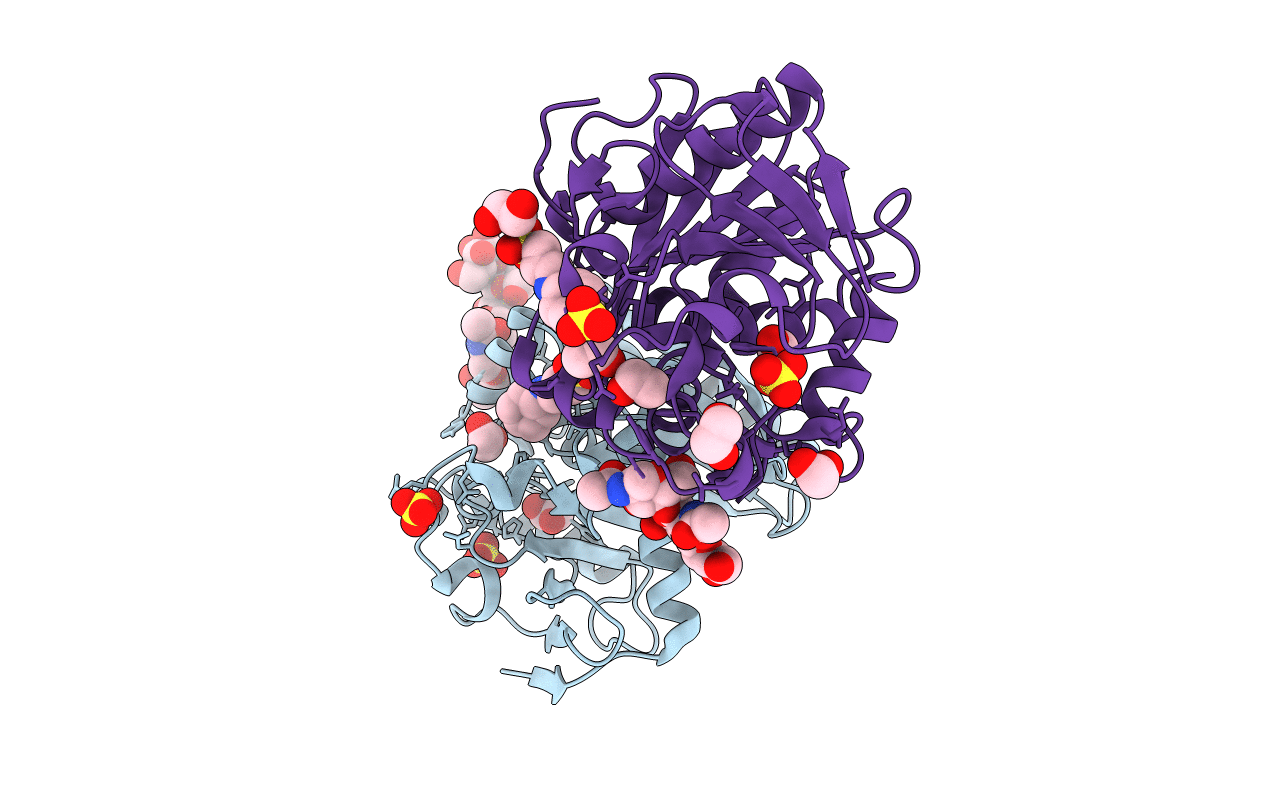 |
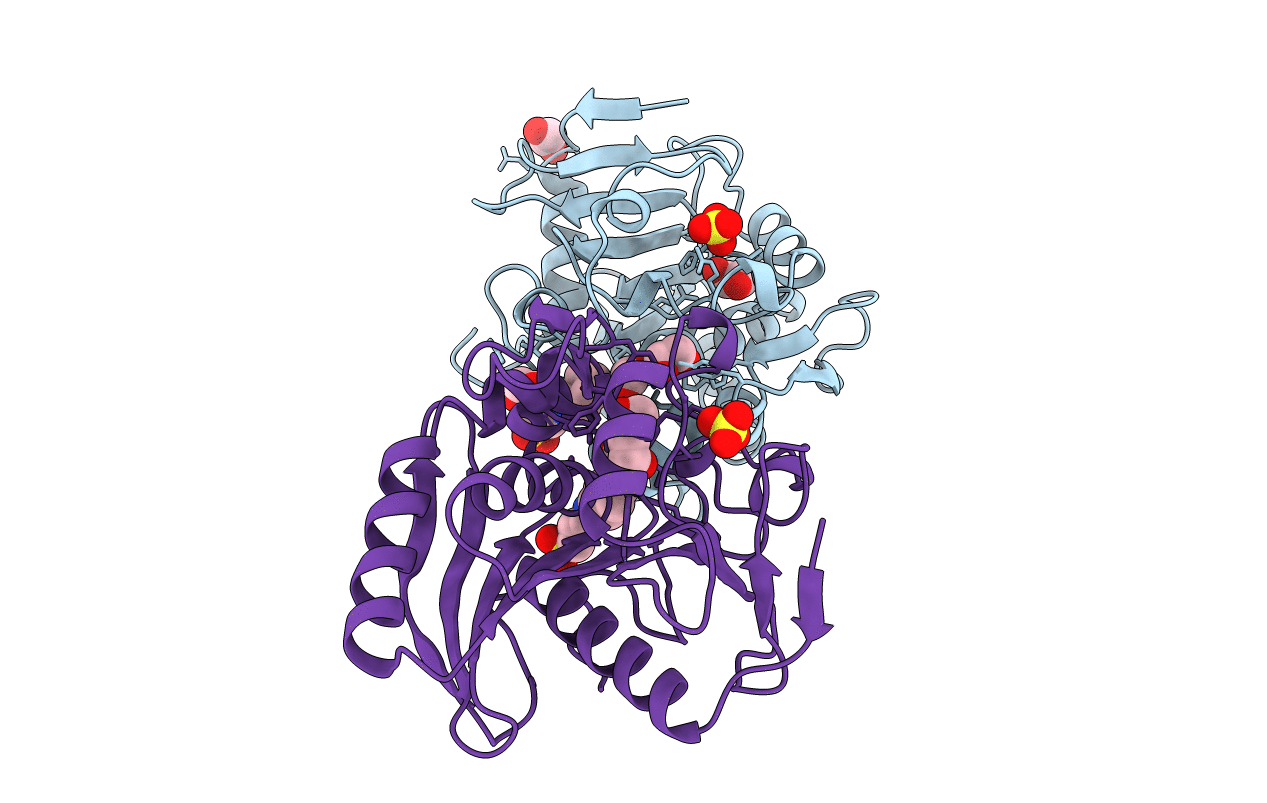
Structure Of Homologously Expressed Ferrulate Esterase Of Aspergillus Niger In Complex With Caps
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2006-10-31 Classification: HYDROLASE Ligands: SO4, EDO, CXS |
 |
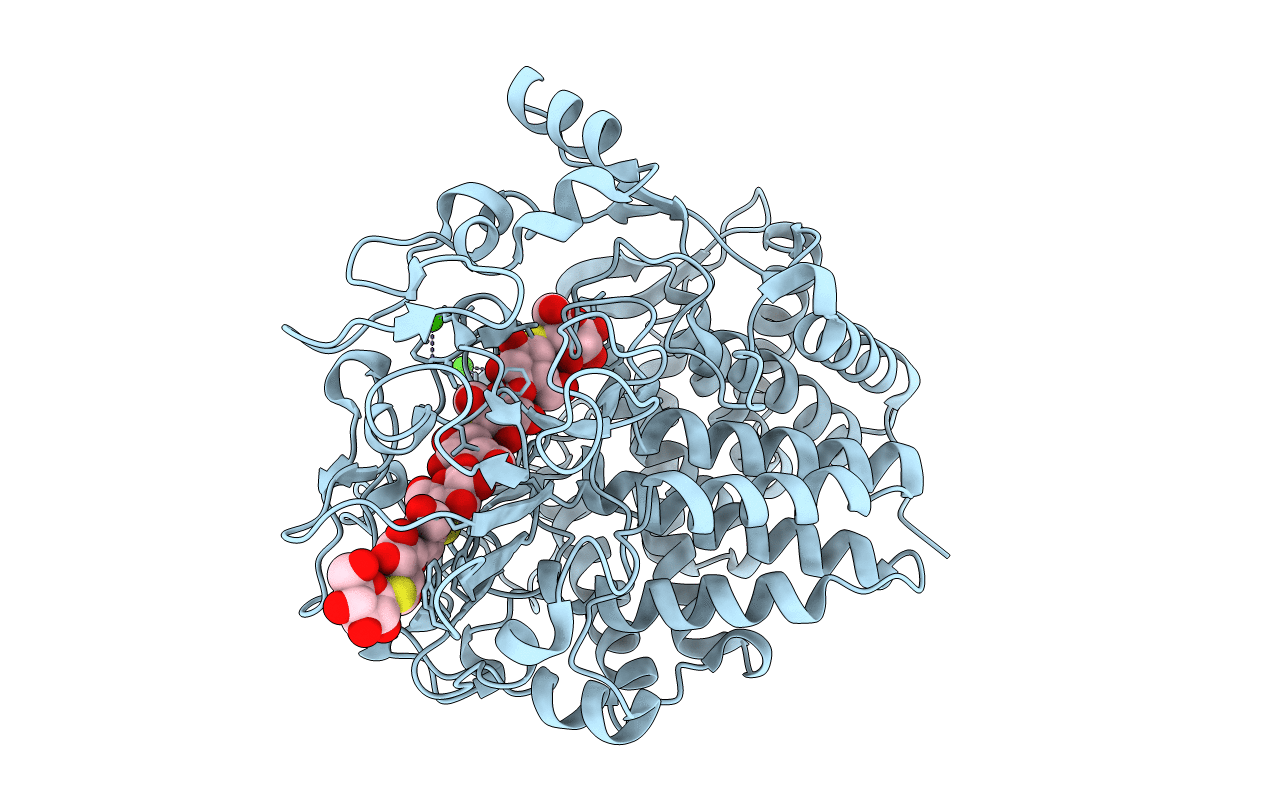
Respective Role Of Protein Folding And Glycosylation In The Thermal Stability Of Recombinant Feruloyl Esterase A
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2006-10-18 Classification: HYDROLASE Ligands: CXS, EDO, SO4 |
 |
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2003-06-24 Classification: HYDROLASE Ligands: CA, MG |
 |
X-Tal Structure Of The Mutant E44Q Of The Cellulase Cel48F In Complex With A Thiooligosaccharide
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2003-06-24 Classification: HYDROLASE Ligands: CA, CL |
 |
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2002-10-30 Classification: HYDROLASE Ligands: CA, ZN, NI, SO4 |
 |
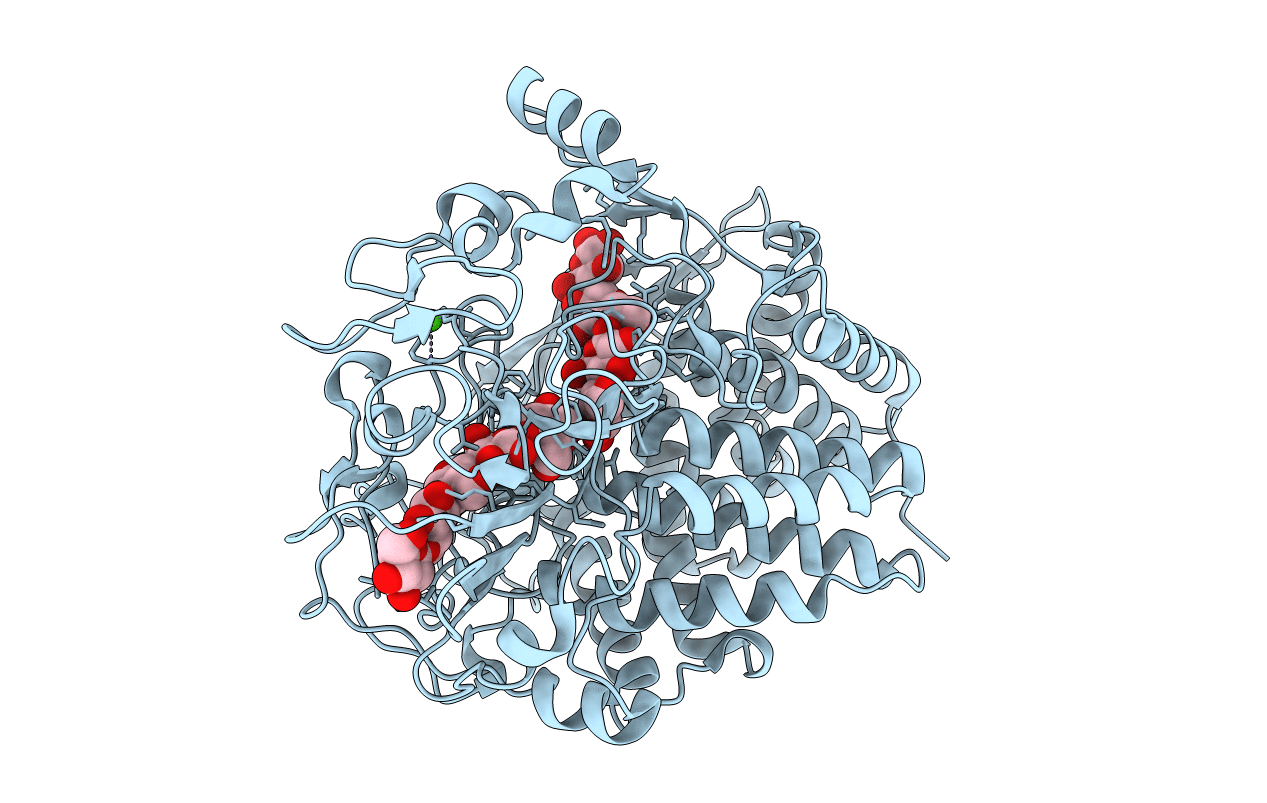
Crystal Structure Of The Cellulase Cel9M Of C. Cellulolyticium In Complex With Cellobiose
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2002-10-30 Classification: HYDROLASE Ligands: CA, ZN, NI, SO4, EDO |
 |
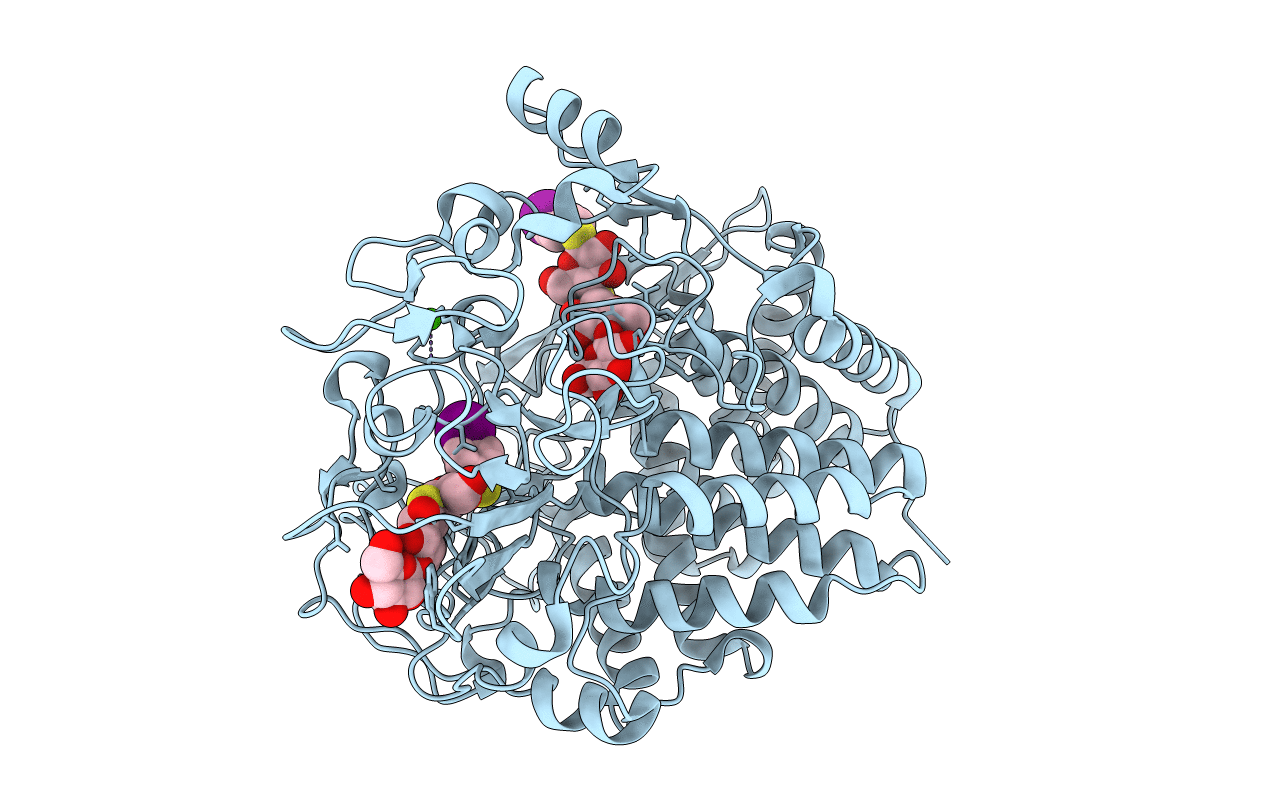
Crystal Structure Of The Cellulase Cel48F From C. Cellulolyticum In Complex With Cellotetraose
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2000-08-02 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of The Cellulase Cel48F From C. Cellulolyticum With The Thiooligosaccharide Inhibitor Pips-Ig3
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2000-08-02 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA |
 |
Crystal Structure Of The Cellulase Cel48F From C. Cellulolyticum In Complex With Cellobiose
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-08-02 Classification: HYDROLASE Ligands: CA |
 |
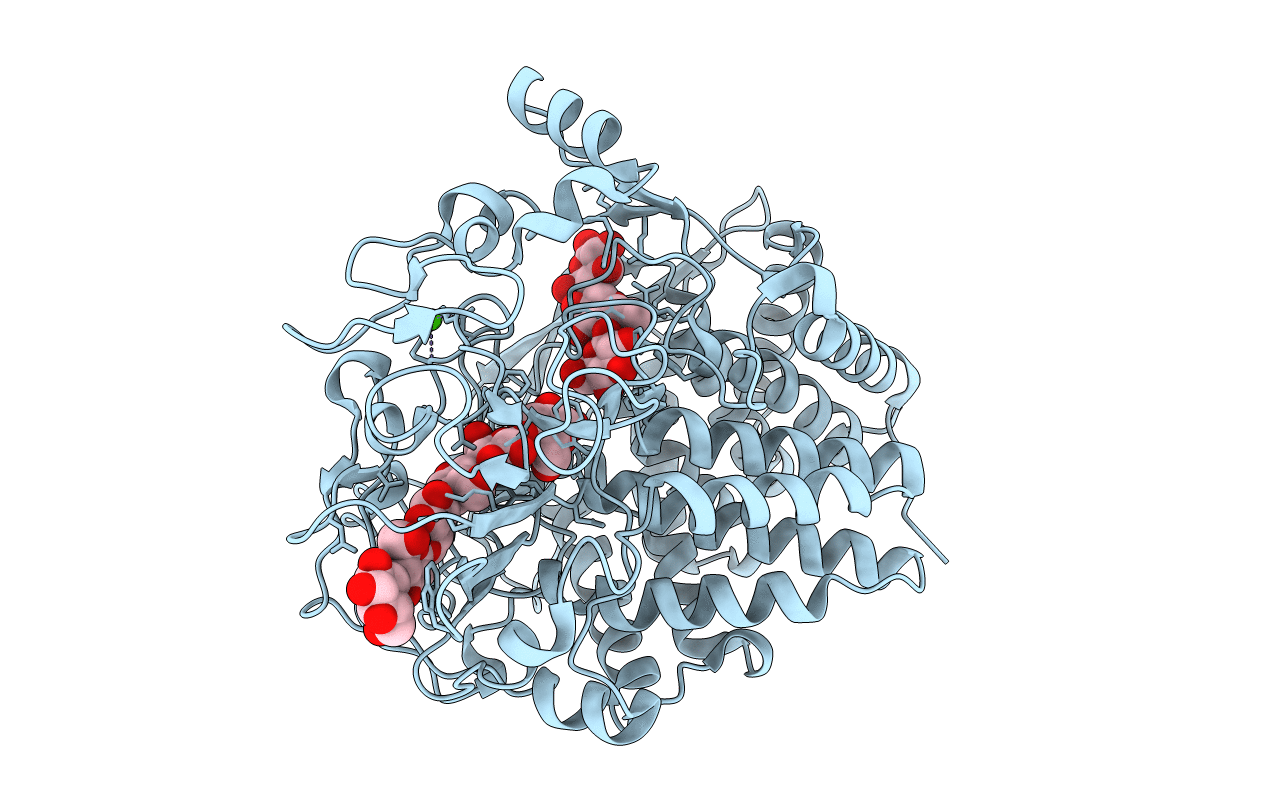
Crystal Structure Of The Cellulase Cel48F From C. Cellulolyticum In Complex With Cellobiitol
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2000-08-02 Classification: HYDROLASE Ligands: CA |
 |
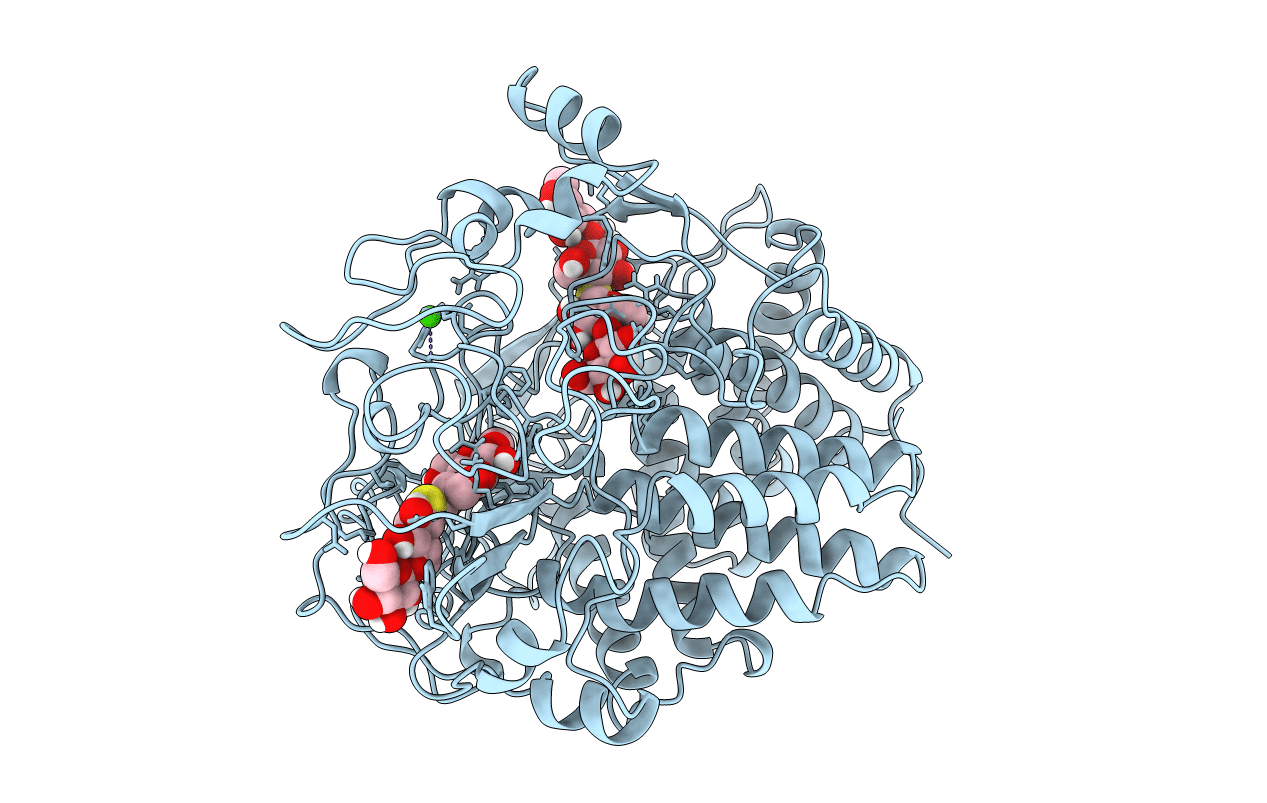
Crystal Structure Of The Cellulase Cel48F From C. Cellulolyticum In Complex With Cellohexaose
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-08-02 Classification: HYDROLASE Ligands: CA |
 |
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1999-07-22 Classification: CELLULASE DEGRADATION Ligands: CA |
 |
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 1998-10-14 Classification: GLYCOSYLTRANSFERASE Ligands: OPG, CA |
 |
Structure Of Cyclodextrin Glycosyltransferase Complexed With A Thio-Maltohexaose
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1998-10-14 Classification: TRANSFERASE Ligands: CA |
 |
Structure Of Cyclodextrin Glycosyltransferase Complexed With A Thio-Maltopentaose
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1998-10-14 Classification: TRANSFERASE Ligands: CA |
 |
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 1998-08-12 Classification: GLYCOSYLTRANSFERASE Ligands: CA |
 |
Maltotriose Complex Of Preconditioned Cyclodextrin Glycosyltransferase Mutant
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1998-08-12 Classification: GLYCOSYLTRANSFERASE Ligands: GLC, CA |
 |
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1998-08-12 Classification: GLYCOSYLTRANSFERASE Ligands: CA |

