Search Count: 18
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-08-09 Classification: ELECTRON TRANSPORT Ligands: HEM, LMR, CL |
 |
Organism: Micromonospora maris (strain dsm 45365 / jcm 31040 / nbrc 109089 / nrrl b-24793 / ab-18-032)
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2022-11-30 Classification: OXIDOREDUCTASE Ligands: HEM, PGE, GOL, PEG, P6G, P33, CL, MG |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2021-10-06 Classification: DE NOVO PROTEIN Ligands: HEM |
 |
Organism: Streptococcus gordonii (strain challis / atcc 35105 / bcrc 15272 / ch1 / dl1 / v288)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-05-26 Classification: CELL ADHESION |
 |
Nmr Structure Of Repeat Domain 13 Of The Fibrillar Adhesin Csha From Streptococcus Gordonii.
Organism: Streptococcus gordonii (strain challis / atcc 35105 / bcrc 15272 / ch1 / dl1 / v288)
Method: SOLUTION NMR Release Date: 2020-04-08 Classification: CELL ADHESION |
 |
Organism: Verrucosispora
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-05-16 Classification: HYDROLASE |
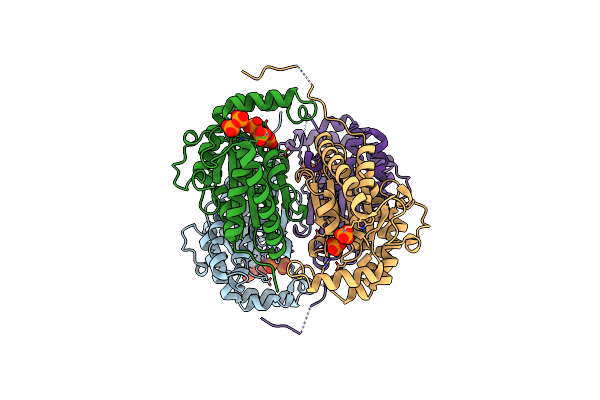 |
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Release Date: 2018-03-21 Classification: TRANSFERASE Ligands: PO4, DPO |
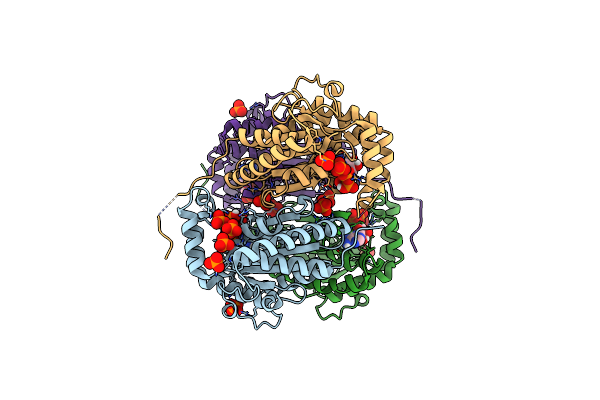 |
Structure Of Polyphosphate Kinase From Meiothermus Ruber N121D Bound To Atp
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-03-21 Classification: TRANSFERASE Ligands: ATP, PO4 |
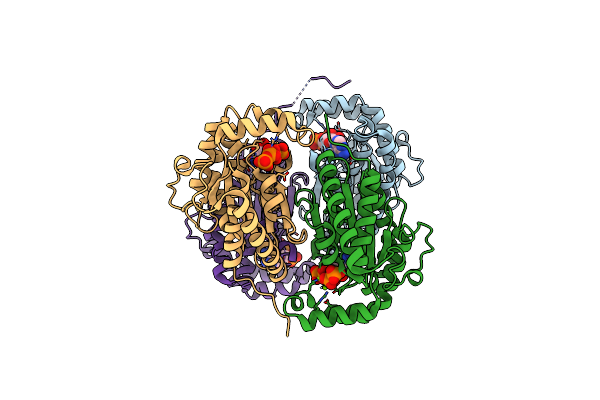 |
Crystal Structure Of Polyphosphate Kinase From Meiothermus Ruber Bound To Adp And Ppi
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2017-12-20 Classification: TRANSFERASE Ligands: ADP, PPV, MG |
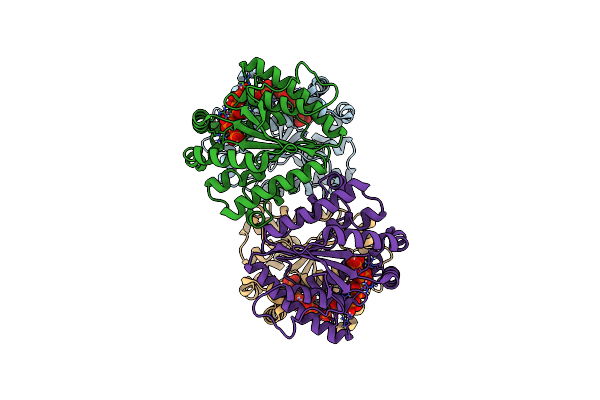 |
Structure Of Polyphosphate Kinase 2 From Francisella Tularensis Schu S4 With Polyphosphate
Organism: Francisella tularensis subsp. tularensis (strain schu s4 / schu 4)
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2017-10-11 Classification: TRANSFERASE Ligands: 9PI |
 |
Structure Of Polyphosphate Kinase 2 From Francisella Tularensis With Amppch2Ppp And Polyphosphate
Organism: Francisella tularensis subsp. tularensis (strain schu s4 / schu 4)
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2017-10-11 Classification: TRANSFERASE Ligands: CL, MG, 6YZ, 6YW |
 |
Structure Of Polyphosphate Kinase 2 Mutant D117N From Francisella Tularensis With Polyphosphate
Organism: Francisella tularensis subsp. tularensis (strain schu s4 / schu 4)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2017-10-11 Classification: TRANSFERASE Ligands: PO4, 6YX, 6YY, CL, 6YW |
 |
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-06-21 Classification: TRANSFERASE Ligands: PO4, SO4 |
 |
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2017-06-21 Classification: TRANSFERASE Ligands: AMP, PO4, SO4, MG |
 |
Crystal Structure Of Polyphosphate Kinase From Meiothermus Ruber Bound To Atp
Organism: Meiothermus ruber
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2017-06-21 Classification: TRANSFERASE Ligands: ATP, MG, SO4, GOL |
 |
Crystal Structure Of Polyphosphate Kinase From Meiothermus Ruber Bound To Adp
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-06-21 Classification: TRANSFERASE Ligands: ADP, PO4, MG, GOL, CL |
 |
Organism: Verrucosispora maris
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-06-29 Classification: HYDROLASE |
 |
Characterisation Of Polyphosphate Kinase 2 From The Intracellular Pathogen Francisella Tularensis
Organism: Francisella tularensis
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2015-12-02 Classification: TRANSFERASE |

