Search Count: 111
 |
The Structure Of 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase With Gly190Pro Mutation
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-04-08 Classification: TRANSFERASE Ligands: PO4, SO4, MN, CL, PEG, PG4 |
 |
Crystal Structure Of Chorismate Mutase From Helicobacter Pylori In Complex With Prephenate
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-11-07 Classification: ISOMERASE Ligands: PRE, PYR, PHB |
 |
The Structure Of A Dimeric Type Ii Dah7Ps Associated With Pyocyanin Biosynthesis In Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-10-03 Classification: TRANSFERASE Ligands: PEP, CO, CL |
 |
Structure Of Bacterial Type Ii Nadh Dehydrogenase From Caldalkalibacillus Thermarum Complexed With A Quinone Inhibitor Hqno At 2.8A Resolution
Organism: Caldalkalibacillus thermarum ta2.a1
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-05-16 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: FAD, HQO |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2018-02-28 Classification: TRANSFERASE Ligands: PEP, TRP, PO4, CO, CL |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-02-28 Classification: TRANSFERASE Ligands: PO4, CO, PEP, TYR, CL |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2018-02-28 Classification: TRANSFERASE Ligands: CO, PEP, CL, ACT |
 |
Catalytic Core Domain Of Adenosine Triphosphate Phosphoribosyltransferase From Campylobacter Jejuni
Organism: Campylobacter jejuni (strain rm1221)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-12-20 Classification: TRANSFERASE Ligands: ZN, ACT, CL |
 |
Catalytic Core Domain Of Adenosine Triphosphate Phosphoribosyltransferase From Campylobacter Jejuni With Bound Phosphoribosyl-Atp
Organism: Campylobacter jejuni (strain rm1221)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-12-20 Classification: TRANSFERASE Ligands: ZN, PRT, CL |
 |
Catalytic Core Domain Of Adenosine Triphosphate Phosphoribosyltransferase From Campylobacter Jejuni With Bound Atp
Organism: Campylobacter jejuni (strain rm1221)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-12-20 Classification: TRANSFERASE Ligands: ZN, ATP, PO4, ACT |
 |
Catalytic Core Domain Of Adenosine Triphosphate Phosphoribosyltransferase From Campylobacter Jejuni With Bound Prpp
Organism: Campylobacter jejuni (strain rm1221)
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2017-12-20 Classification: TRANSFERASE Ligands: ZN, ACT, PRP, MG, EDO |
 |
Structure Of Bacterial Type Ii Nadh Dehydrogenase From Caldalkalibacillus Thermarum At 2.15A Resolution
Organism: Caldalkalibacillus thermarum ta2.a1
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-10-18 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Transition State Analysis Of Adenosine Triphosphate Phosphoribosyltransferase
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-09-20 Classification: TRANSFERASE Ligands: SO4, MG, ATP |
 |
Crystal Structure Of Dah7Ps-Cm Complex From Geobacillus Sp. With Prephenate
Organism: Geobacillus sp. ghh01
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2016-09-07 Classification: TRANSFERASE,ISOMERASE Ligands: MN, SO4, PRE |
 |
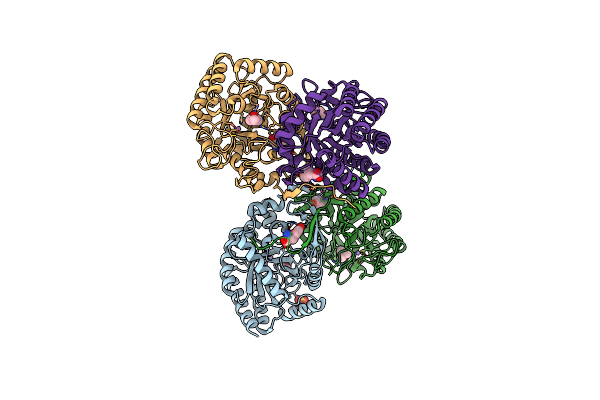
Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase Regulated And Complexed With Pep
Organism: Neisseria meningitidis serogroup b (strain mc58)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2016-08-24 Classification: TRANSFERASE Ligands: MN, SO4, EDO, PEG, PGE, PEP, PHE, CL |
 |
Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase Regulated (Tyrosine)
Organism: Neisseria meningitidis serogroup b (strain mc58)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2016-08-24 Classification: TRANSFERASE Ligands: MN, SO4, TMO, TYR |
 |
Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase Regulated (Tryptophan)
Organism: Neisseria meningitidis serogroup b (strain mc58)
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2016-08-24 Classification: TRANSFERASE Ligands: MN, TRP |
 |
Neisseria Meningitidis 3 Dexy-D-Arabino-Heptulosonate 7-Phosphate Synthase Glu98Ala Variant
Organism: Neisseria meningitidis serogroup b (strain mc58)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-08-10 Classification: TRANSFERASE Ligands: MN, SO4, PEP, EDO |
 |
Neisseria Meningitidis 3 Dexy-D-Arabino-Heptulosonate 7-Phosphate Synthase Glu98Ala Variant Regulated
Organism: Neisseria meningitidis serogroup b (strain mc58)
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2016-08-10 Classification: TRANSFERASE Ligands: MN, PEP, PHE, EDO, PEG |
 |
Neisseria Meningitidis 3 Dexy-D-Arabino-Heptulosonate 7-Phosphate Synthase Glu176Ala Variant
Organism: Neisseria meningitidis serogroup b (strain mc58)
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2016-08-10 Classification: TRANSFERASE Ligands: MN, PHE, CL |

