Search Count: 173
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PEPTIDE BINDING PROTEIN Ligands: EDO, IMD, PEG |
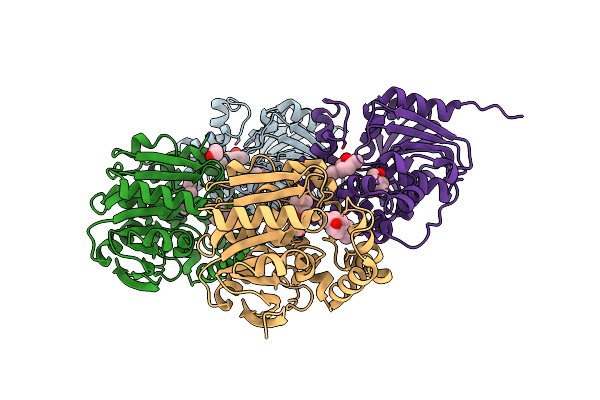 |
Limonene Epoxide Low Ph Soak Of Epoxide Hydrolase From Metagenomic Source Ch65
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: LEO, CL, EDO |
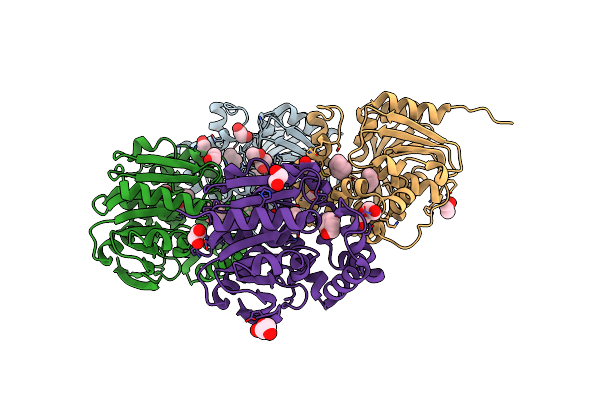 |
Cyclohexane Epoxide Low Ph Soak Of Epoxide Hydrolase From Metagenomic Source Ch65
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: 7EX, PEG, EDO |
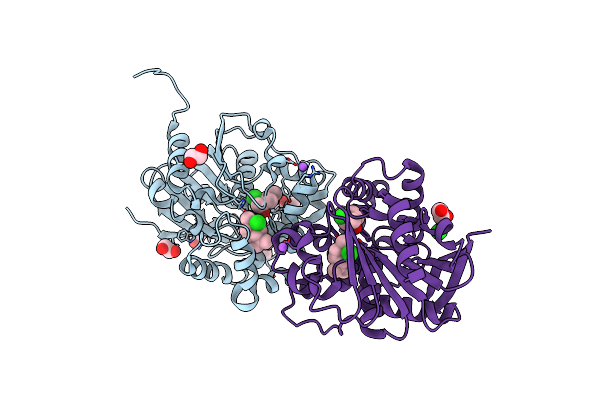 |
Halogenated Product Of Limonene Epoxide Turnover By Epoxide Hydrolase From Metagenomic Source Ch65
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: M1U, CL, EDO, NA |
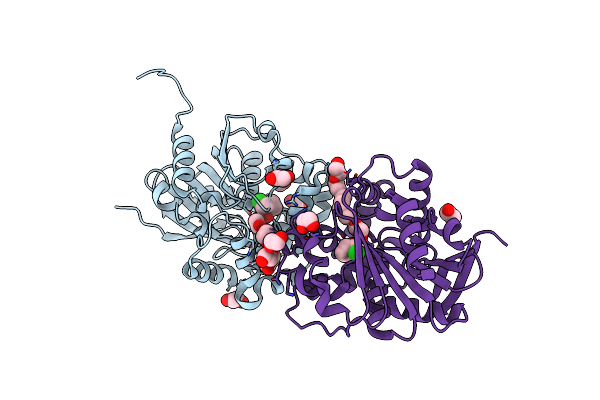 |
Cyclohexane Epoxide Soak Of Epoxide Hydrolase From Metagenomic Source Ch65 Resulting In Halogenated Compound In The Active Site
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: 2CH, EDO, PGE, CL, P6G |
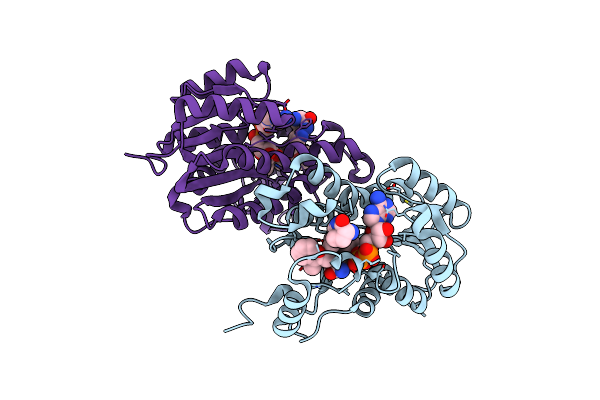 |
Crystal Structure Of E. Coli Fabi In Complex With Nad And (R,E)-3-(7-Amino-8-Oxo-6,7,8,9-Tetrahydro-5H-Pyrido[2,3-B]Azepin-3-Yl)-N-Methyl-N-((3-Methylbenzofuran-2-Yl)Methyl)Acrylamide
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-02-15 Classification: ANTIBIOTIC Ligands: NAD, NQF |
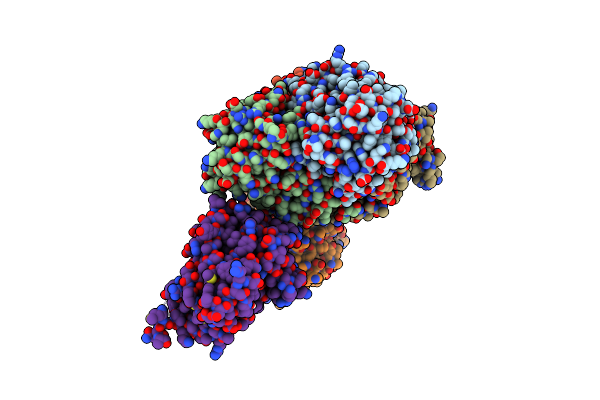 |
Crystal Structure Of Acinetobacter Baumannii Fabi In Complex With Nad And (R,E)-3-(7-Amino-8-Oxo-6,7,8,9-Tetrahydro-5H-Pyrido[2,3-B]Azepin-3-Yl)-N-Methyl-N-((3-Methylbenzofuran-2-Yl)Methyl)Acrylamide
Organism: Acinetobacter baumannii atcc 19606 = cip 70.34 = jcm 6841
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-02-15 Classification: ANTIBIOTIC Ligands: NQF, NAD |
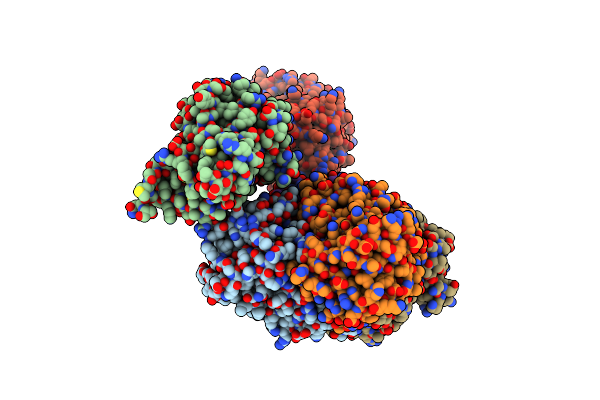 |
Crystal Structure Of Acinetobacter Baumannii Fabi In Complex With Nad And Fabimycin ((S,E)-3-(7-Amino-8-Oxo-6,7,8,9-Tetrahydro-5H-Pyrido[2,3-B]Azepin-3-Yl)-N-Methyl-N-((3-Methylbenzofuran-2-Yl)Methyl)Acrylamide)
Organism: Acinetobacter baumannii atcc 19606 = cip 70.34 = jcm 6841
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2023-02-15 Classification: ANTIBIOTIC Ligands: NAD, NUC |
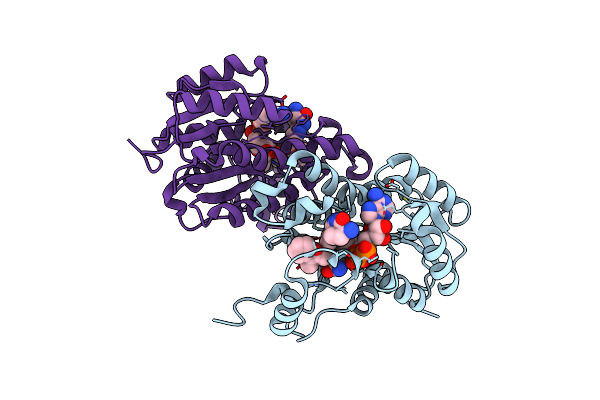 |
Crystal Structure Of E. Coli Fabi In Complex With Nad And Fabimycin ((S,E)-3-(7-Amino-8-Oxo-6,7,8,9-Tetrahydro-5H-Pyrido[2,3-B]Azepin-3-Yl)-N-Methyl-N-((3-Methylbenzofuran-2-Yl)Methyl)Acrylamide)
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2022-12-14 Classification: ANTIBIOTIC Ligands: NAD, NUC |
 |
Crystal Structure Of Sars-Cov-2 Spike Protein N-Terminal Domain In Complex With Biliverdin
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2021-04-28 Classification: VIRAL PROTEIN Ligands: BLA, NAG, PG4, PEG, PGE, 1PE |
 |
Trimeric Sars-Cov-2 Spike Ectodomain In Complex With Biliverdin (Closed Conformation)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2021-04-28 Classification: VIRAL PROTEIN Ligands: BLA, NAG |
 |
Trimeric Sars-Cov-2 Spike Ectodomain In Complex With Biliverdin (One Rbd Erect)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2021-04-28 Classification: VIRAL PROTEIN Ligands: BLA, NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-04-28 Classification: VIRAL PROTEIN Ligands: BLA, NAG |
 |
The Structure Of 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase With Gly190Pro Mutation
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-04-08 Classification: TRANSFERASE Ligands: PO4, SO4, MN, CL, PEG, PG4 |
 |
Crystal Structure Determination Of Human/Porcine Chimera Coagulation Factor Viii
Organism: Sus scrofa, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2019-09-11 Classification: BLOOD CLOTTING Ligands: CA, ZN, CU1 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.61 Å Release Date: 2019-09-11 Classification: BLOOD CLOTTING Ligands: CA, ZN, CU1 |
 |
Crystal Structure Of Chorismate Mutase From Helicobacter Pylori In Complex With Prephenate
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-11-07 Classification: ISOMERASE Ligands: PRE, PYR, PHB |
 |
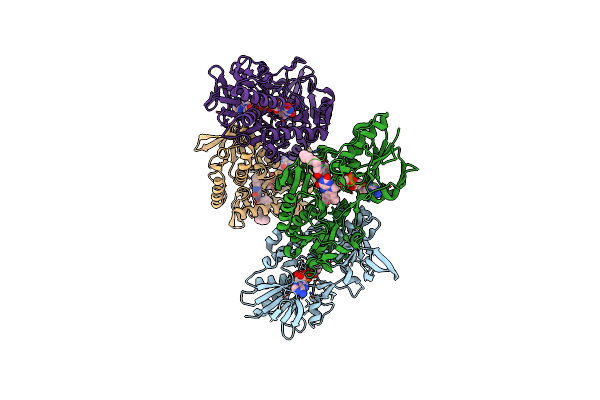
The Structure Of A Dimeric Type Ii Dah7Ps Associated With Pyocyanin Biosynthesis In Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-10-03 Classification: TRANSFERASE Ligands: PEP, CO, CL |
 |
Structure Of Bacterial Type Ii Nadh Dehydrogenase From Caldalkalibacillus Thermarum Complexed With A Quinone Inhibitor Hqno At 2.8A Resolution
Organism: Caldalkalibacillus thermarum ta2.a1
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-05-16 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: FAD, HQO |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2018-02-28 Classification: TRANSFERASE Ligands: PEP, TRP, PO4, CO, CL |

