Search Count: 86
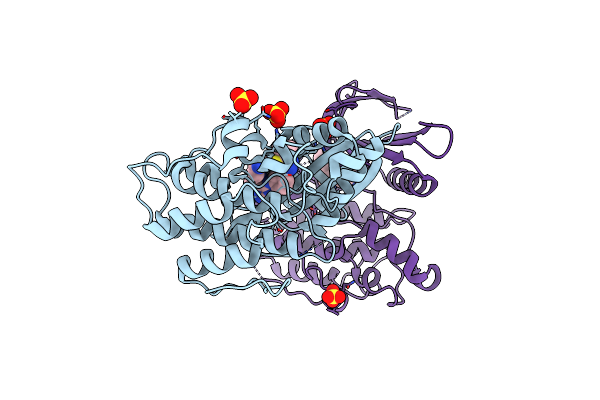 |
Crystal Structure Of Ap2 Associated Kinase 1 Complexed With 5-[(4-Aminopiperidin-1-Yl)Methyl]-N-{3-[5-(Propan-2-Yl)-1,3,4-Thiadiazol-2-Yl]Phenyl}Pyrrolo[2,1-F][1,2,4]Triazin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-07-12 Classification: TRANSFERASE Ligands: YFV, SO4 |
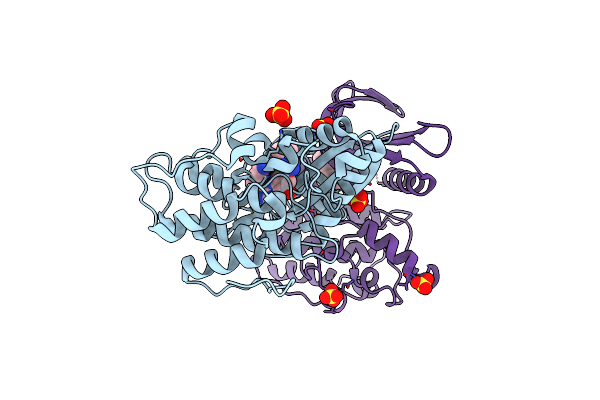 |
Crystal Structure Of Ap2 Associated Kinase 1 Complexed With (5P)-3-({(8R)-5-[(4-Aminopiperidin-1-Yl)Methyl]Pyrrolo[2,1-F][1,2,4]Triazin-4-Yl}Amino)-5-[2-(Propan-2-Yl)-2H-Tetrazol-5-Yl]Phenol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-07-12 Classification: TRANSFERASE Ligands: ZRR, SO4 |
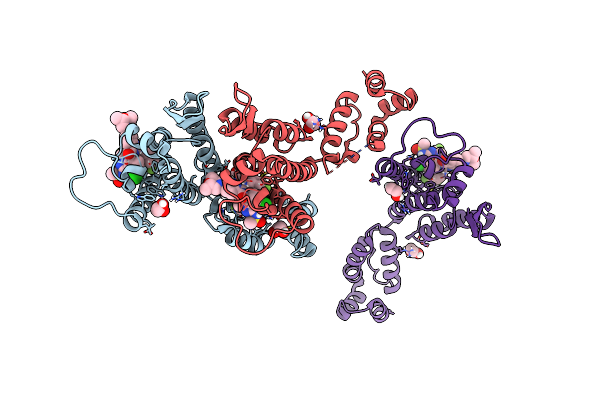 |
Potent Long-Acting Inhibitors Targeting Hiv-1 Capsid Based On A Versatile Quinazolin-4-One Scaffold
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2023-02-15 Classification: ANTIVIRAL-PROTEIN/INHIBITOR Ligands: EDO, Y05 |
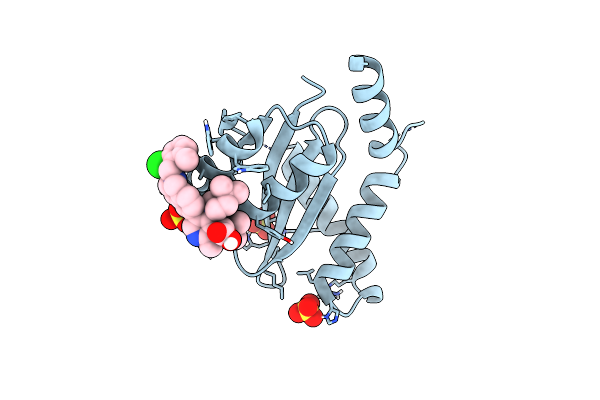 |
Crystal Structure Of Hiv-1 Integrase Complexed With (2S)-2-(Tert-Butoxy)-2-(5-{2-[(2-Chloro-6-M Ethylphenyl)Methyl]-1,2,3,4-Tetrahydroisoquinolin-6-Yl}-4- (4,4-Dimethylpiperidin-1-Yl)-2-Methylpyridin-3-Yl)Acetic Acid
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: O20, SO4 |
 |
Crystal Structure Of Hiv-1 Integrase Complexed With Compound-2A Aka (2S)-2-(Tert-Butoxy)-2-[7-(4,4-Dimethylpipe Ridin-1-Yl)-8-{4-[2-(4-Fluorophenyl)Ethoxy]Phenyl}-2,5-Dim Ethylimidazo[1,2-A]Pyridin-6-Yl]Acetic Acid
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-06-08 Classification: DNA BINDING PROTEIN, viral protein Ligands: KZD, SO4 |
 |
Organism: human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2022-04-06 Classification: DNA BINDING PROTEIN, Viral protein Ligands: MG, GE7 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-04-06 Classification: DNA BINDING PROTEIN, Viral Protein Ligands: SO4, GEI, GOL |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, FIV, PEG |
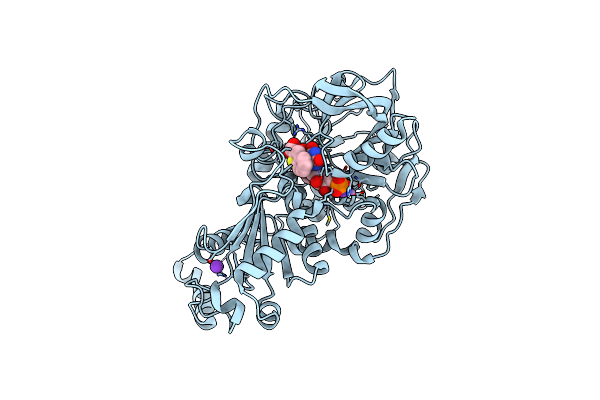 |
Structure Of A. Niger Fdc I327S Variant In Complex With Benzothiophene 2 Carboxylic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, ND8 |
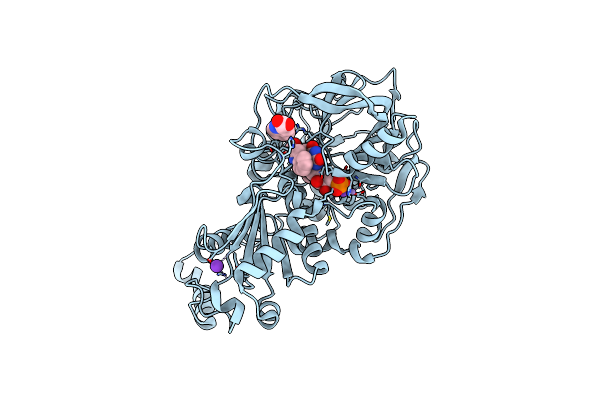 |
Structure Of A. Niger Fdc I327S Variant In Complex With Indol-2-Carboxylic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, ICB |
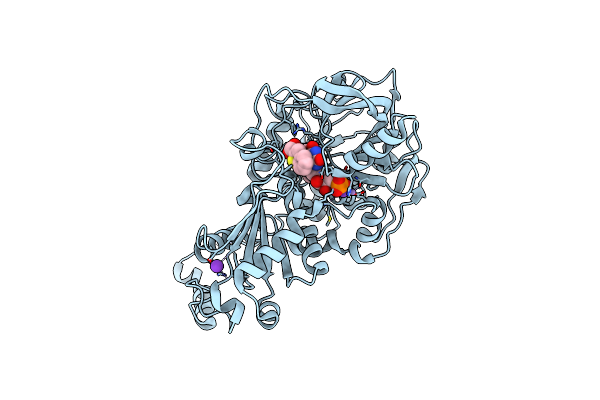 |
Structure Of A. Niger Fdc Wt In Complex With Benzothiophene 2 Carboxylic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, ND8 |
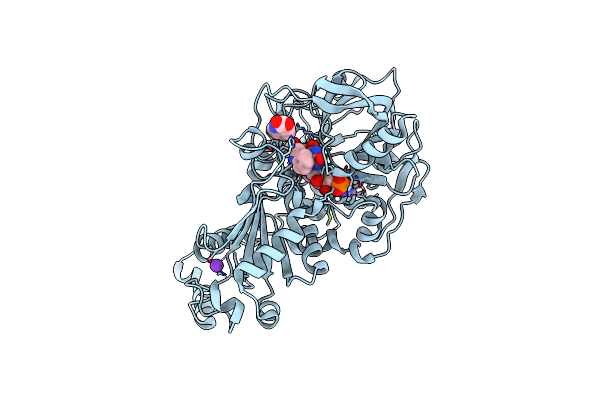 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, ICB |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, FMN, FIV |
 |
Structure Of A. Niger Fdc Wt In Complex With Fmn And Indole 2 Carboxylic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, FMN, ICB |
 |
Structure Of A. Niger Fdc Wt In Complex With Fmn And Benzothiophene 2 Carboxylic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, FMN, ND8 |
 |
Structure Of The Ubid-Class Enzyme Hmff From Pelotomaculum Thermopropionicum In Complex With Fmn
Organism: Pelotomaculum thermopropionicum si
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2019-02-27 Classification: FLAVOPROTEIN Ligands: FMN, K, MN, CA |
 |
Structure Of An Evolved Dimeric Form Of The Ubid-Class Enzyme Hmff From Pelotomaculum Thermopropionicum In Complex With Prfmn
Organism: Pelotomaculum thermopropionicum (strain dsm 13744 / jcm 10971 / si)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-02-27 Classification: FLAVOPROTEIN Ligands: K, MN, 4LU, CA |
 |
Structure Of Hiv-1 Integrase With Potent 5,6,7,8-Tetrahydro-1,6-Naphthyridine Derivatives Allosteric Site Inhibitors
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-01-16 Classification: VIRAL PROTEIN Ligands: EDO, KJJ, SO4 |
 |
Crystal Structure Of The Hepatitis C Virus Genotype 2A Strain Jfh1 Ns5B Rna-Dependent Rna Polymerase In Complex With 6-[Ethyl(Methylsulfonyl)Amino]-2-(4-Fluorophenyl)-N-Methyl-5-(3-{[1-(Pyrimidin-2-Yl)Cyclopropyl]Carbamoyl}Phenyl)-1-Benzofuran-3-Carboxamide
Organism: Hepacivirus c
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2018-11-21 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: J6D, SCN, 2PE, GOL |
 |
Crystal Structure Of The Hepatitis C Virus Genotype 2A Strain Jfh1 L30S Ns5B Rna-Dependent Rna Polymerase In Complex With 6-(Ethylamino)-2-(4-Fluorophenyl)-5-(3-{[1-(5-Fluoropyrimidin-2-Yl)Cyclopropyl]Carbamoyl}-4-Methoxyphenyl)-N-Methyl-1-Benzofuran-3-Carboxamide
Organism: Hepacivirus c
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2018-11-21 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: J6J, SO4, GOL, PG4 |

