Search Count: 17
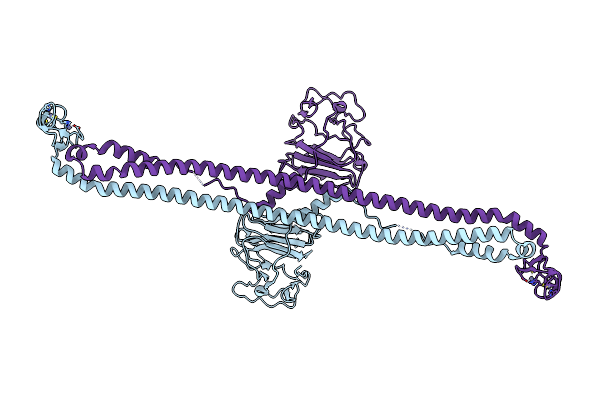 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:7.10 Å Release Date: 2023-07-12 Classification: MEMBRANE PROTEIN Ligands: ZN |
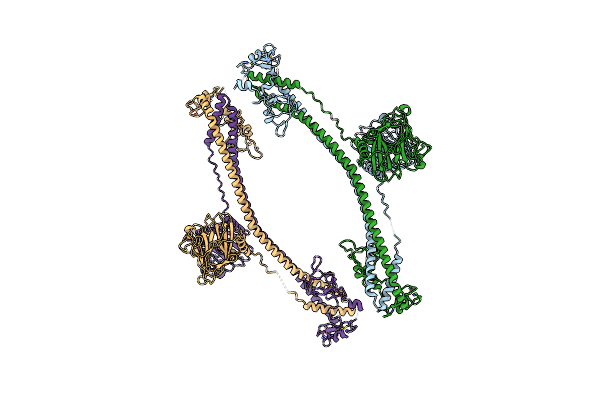 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:4.62 Å Release Date: 2023-07-12 Classification: MEMBRANE PROTEIN Ligands: ZN |
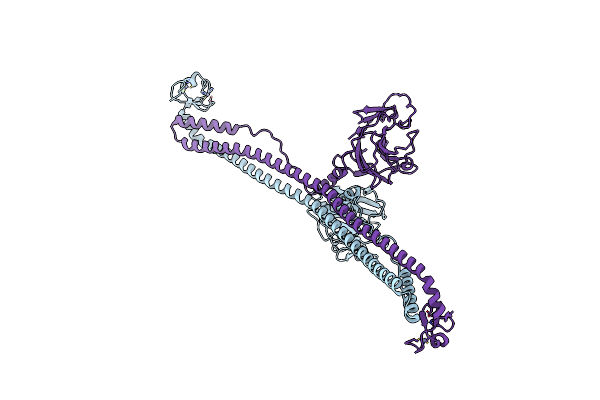 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.28 Å Release Date: 2023-07-12 Classification: MEMBRANE PROTEIN Ligands: ZN |
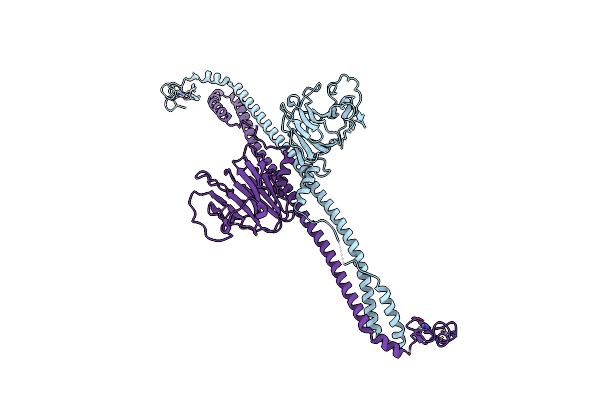 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:5.20 Å Release Date: 2023-07-12 Classification: MEMBRANE PROTEIN Ligands: ZN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2023-07-12 Classification: MEMBRANE PROTEIN Ligands: ZN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2023-05-24 Classification: MEMBRANE PROTEIN Ligands: ZN |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2021-04-28 Classification: PEPTIDE BINDING PROTEIN Ligands: ACY, PHE, ALA |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-12-04 Classification: TRANSFERASE Ligands: TMP, IOD, NA |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-11-27 Classification: PROTEIN BINDING/PROTEIN TRANSPORT |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2019-11-27 Classification: PROTEIN BINDING/PROTEIN TRANSPORT |
 |
Organism: Legionella pneumophila subsp. pneumophila str. philadelphia 1
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-08-29 Classification: HYDROLASE Ligands: NCA |
 |
Organism: Legionella pneumophila subsp. pneumophila
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2018-08-29 Classification: HYDROLASE |
 |
Organism: Legionella pneumophila subsp. pneumophila
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-08-29 Classification: HYDROLASE |
 |
 |
Crystal Structure Of The N-Terminal Domain Of The Human Mitochondrial Calcium Uniporter Fused With T4 Lysozyme
Organism: Enterobacteria phage t4, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-09-16 Classification: TRANSPORT PROTEIN Ligands: SO4 |
 |
Crystal Structure Of The N-Terminal Domain Of The Human Mitochondrial Calcium Uniporter
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-09-16 Classification: TRANSPORT PROTEIN Ligands: PG4 |
 |
Crystal Structure Of The N-Terminal Domain Single Mutant (S92A) Of The Human Mitochondrial Calcium Uniporter Fused With T4 Lysozyme
Organism: Enterobacteria phage t4, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2015-09-16 Classification: TRANSPORT PROTEIN Ligands: SO4 |

