Search Count: 27
 |
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: CHAPERONE Ligands: GTP, MG |
 |
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: CHAPERONE Ligands: GTP, MG |
 |
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: CHAPERONE Ligands: GTP, MG |
 |
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: CHAPERONE Ligands: GTP, MG |
 |
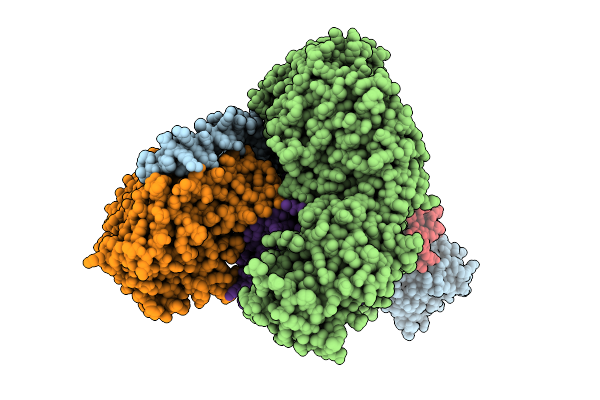
Cryo-Em Structure Of The Tbc-Dec-Arl2-Alpha-Beta-Tubulin Complex With Gdp-Alfx
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: CHAPERONE Ligands: GDP, AF3, MG, GTP |
 |
Cryo-Em Structure Of The Tbc-Dc-Arl2-Alpha-Beta-Tubulin Complex With Gdp-Alfx
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: CHAPERONE Ligands: GDP, AF3, MG, GTP |
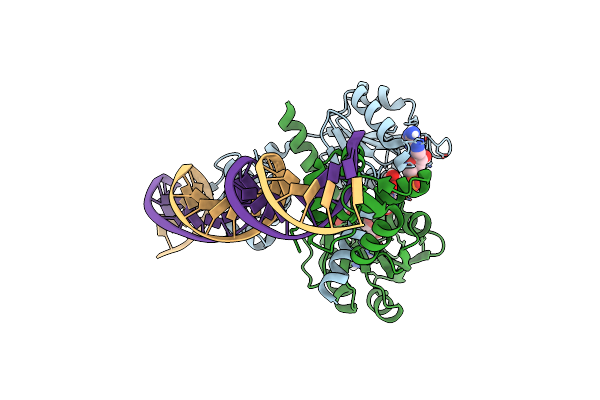 |
Organism: Thermotoga maritima msb8, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-02-16 Classification: GENE REGULATION/DNA Ligands: NAD |
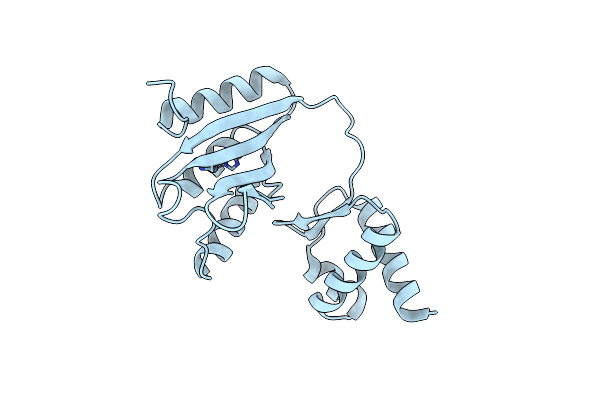 |
Organism: Bacillus halodurans (strain atcc baa-125 / dsm 18197 / ferm 7344 / jcm 9153 / c-125)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-12-16 Classification: STRUCTURAL PROTEIN Ligands: ZN |
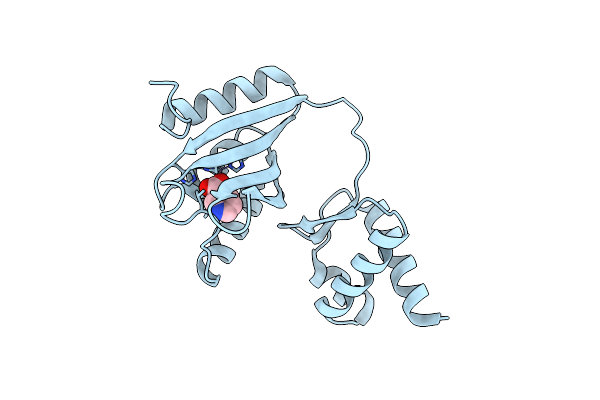 |
Organism: Bacillus halodurans (strain atcc baa-125 / dsm 18197 / ferm 7344 / jcm 9153 / c-125)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-12-16 Classification: STRUCTURAL PROTEIN Ligands: NIO, ZN |
 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-11-07 Classification: GENE REGULATION Ligands: GOL |
 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-11-07 Classification: GENE REGULATION Ligands: NAD, ADP |
 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2018-11-07 Classification: GENE REGULATION Ligands: NAI, GOL |
 |
Organism: Helicobacter pylori (strain atcc 700392 / 26695)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-08-29 Classification: LIGASE |
 |
Organism: Helicobacter pylori (strain atcc 700392 / 26695)
Method: X-RAY DIFFRACTION Resolution:2.93 Å Release Date: 2018-08-29 Classification: LIGASE Ligands: ATP, ADP, PPQ, MG, P3P |
 |
Structural Analysis Of Fatty Acid Degradation Regulator Fadr From Bacillus Halodurans
Organism: Bacillus halodurans (strain atcc baa-125 / dsm 18197 / ferm 7344 / jcm 9153 / c-125)
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2017-03-01 Classification: TRANSCRIPTION, DNA BINDING PROTEIN Ligands: MG, GOL, PLM |
 |
Structural Analysis Of Fatty Acid Degradation Regulator Fadr From Bacillus Halodurans
Organism: Bacillus halodurans (strain atcc baa-125 / dsm 18197 / ferm 7344 / jcm 9153 / c-125)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2017-03-01 Classification: TRANSCRIPTION, DNA BINDING PROTEIN Ligands: MG, GOL |
 |
Structural Analysis Of Fatty Acid Degradation Regulator Fadr From Bacillus Halodurans
Organism: Bacillus halodurans, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-03-01 Classification: TRANSCRIPTION, DNA BINDING PROTEIN/DNA |
 |
Organism: Human herpesvirus 8
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-01-18 Classification: TRANSFERASE Ligands: PO4 |
 |
Organism: Human herpesvirus 8
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-01-18 Classification: TRANSFERASE Ligands: UMP |
 |
Organism: Human herpesvirus 8
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-01-18 Classification: TRANSFERASE Ligands: D16, UMP |

