Search Count: 23
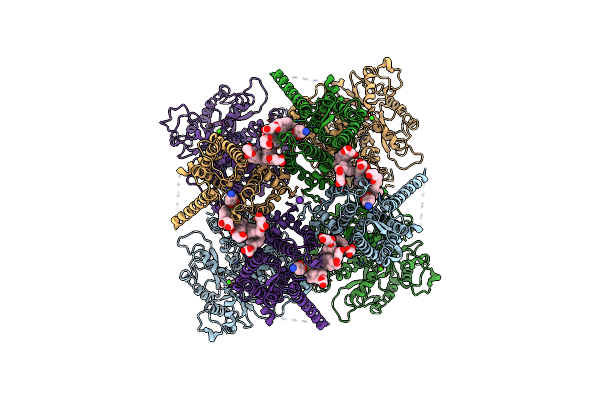 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: MEMBRANE PROTEIN Ligands: MG, CA, K, A1L4E, Y01 |
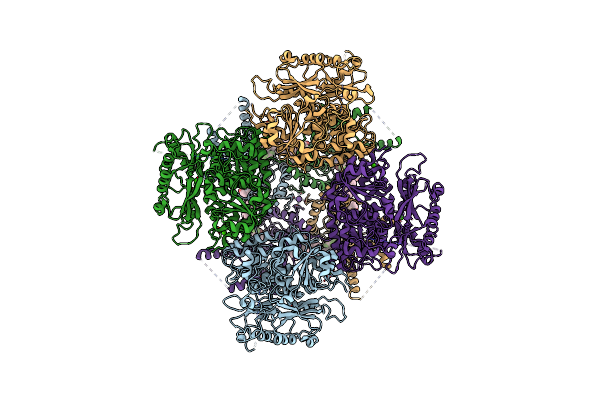 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: MEMBRANE PROTEIN Ligands: A1L4F, MG, CA, K, Y01 |
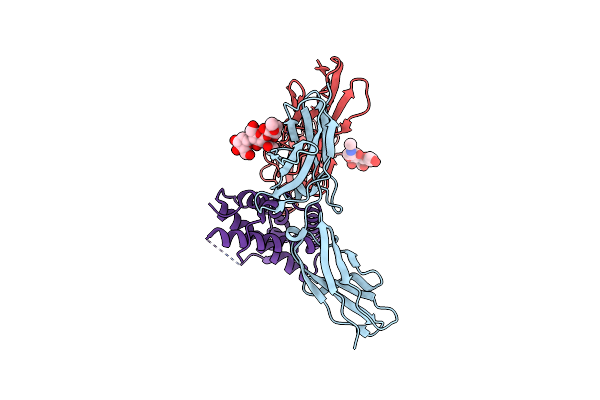 |
Structure Of The Ifn-Lambda4/Ifn-Lambdar1/Il-10Rbeta Receptor Complex With An Engineered Il-10Rbeta
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: CYTOKINE |
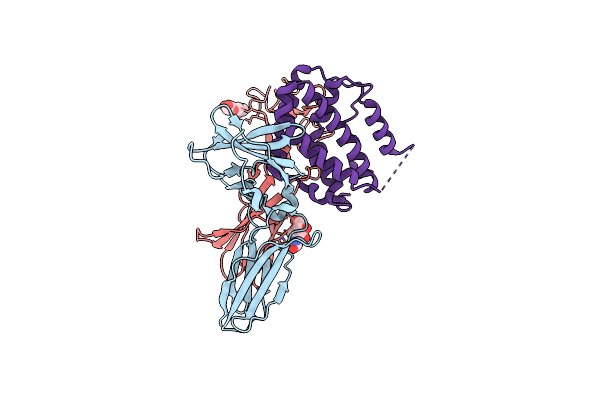 |
Structure Of The Ifn-Lambda3/Ifn-Lambdar1/Il-10Rbeta Receptor Complex With An Engineered Il-10Rbeta
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: CYTOKINE Ligands: NAG |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2021-11-10 Classification: LIPID BINDING PROTEIN Ligands: V8P |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-11-10 Classification: LIPID BINDING PROTEIN/IMMUNE SYSTEM Ligands: QOD, HP6 |
 |
Organism: Epsilonproteobacteria bacterium (ex lamellibrachia satsuma)
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2021-10-27 Classification: HYDROLASE Ligands: MN |
 |
Crystal Structure Of The Extended-Spectrum Class C Beta-Lactamase Ampc Ber With The Ordered R2 Loop
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2021-05-19 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2019-10-16 Classification: TRANSCRIPTION Ligands: A9C |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-10-16 Classification: TRANSCRIPTION Ligands: A9F |
 |
The Ligand-Free Structure Of Human Ppargamma Lbd In The Presence Of The Src-1 Coactivator Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2019-10-16 Classification: TRANSCRIPTION |
 |
Organism: Vibrio vulnificus mo6-24/o
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2019-04-03 Classification: OXIDOREDUCTASE Ligands: MES, EDO |
 |
Organism: Vibrio vulnificus mo6-24/o
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-04-03 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
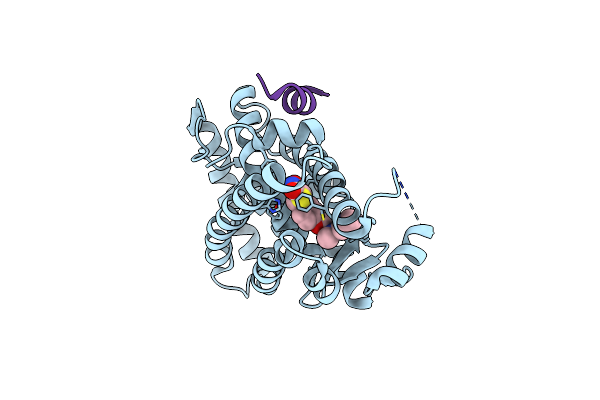
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-09-12 Classification: TRANSCRIPTION Ligands: 8LX |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-09-12 Classification: TRANSCRIPTION Ligands: BRL |
 |
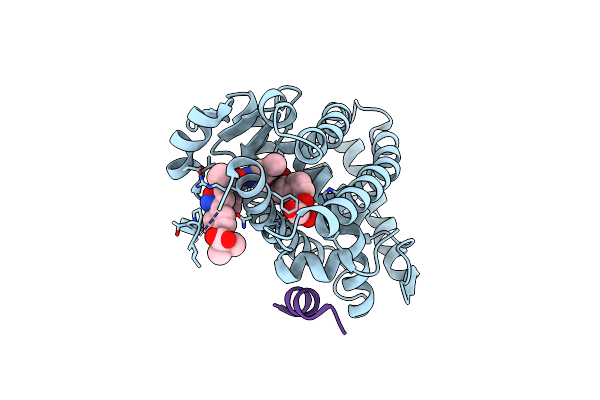
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-07-05 Classification: TRANSCRIPTION Ligands: Q35 |
 |
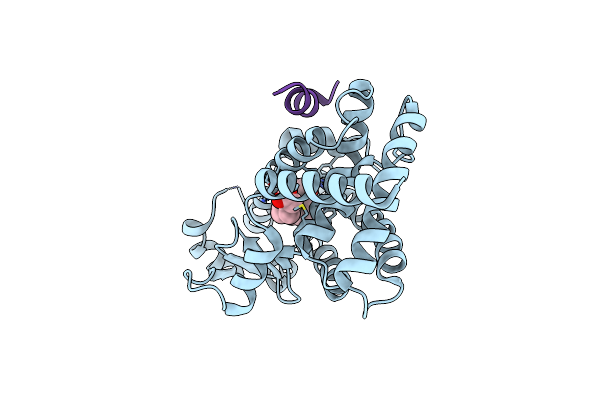
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-07-05 Classification: TRANSCRIPTION Ligands: T35, MYR, GOL |
 |
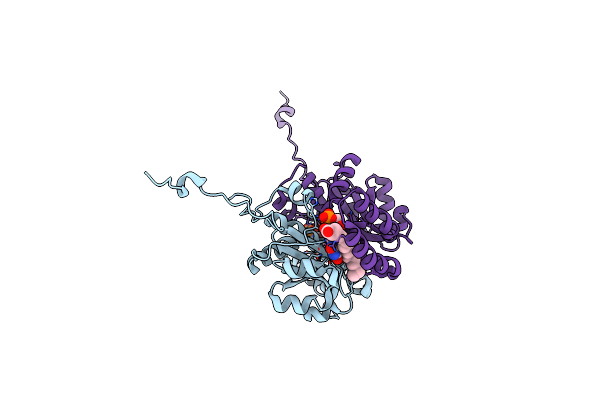
The Agonist-Free Structure Of Human Ppargamma Ligand Binding Domain In The Presence Of The Src-1 Coactivator Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2017-07-05 Classification: TRANSCRIPTION Ligands: MYR, GOL |
 |
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-02-25 Classification: OXIDOREDUCTASE Ligands: FMN, KTC, CL |
 |
Crystal Structure Of Cla-Er, A Novel Enone Reductase Catalyzing A Key Step Of A Gut-Bacterial Fatty Acid Saturation Metabolism, Biohydrogenation
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2015-02-25 Classification: OXIDOREDUCTASE Ligands: FMN |

