Search Count: 44
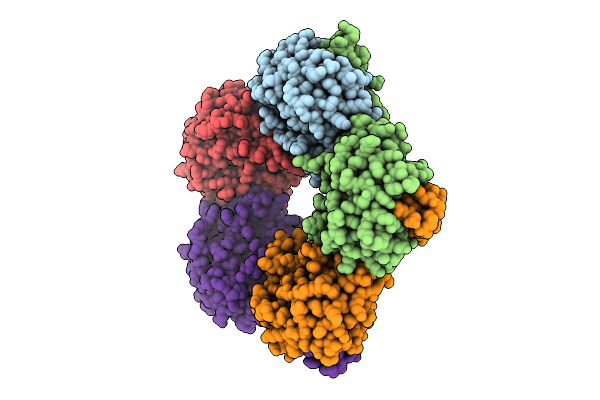 |
Selective Production Of Versatile L-Glyceraldehyde From C1 And/Or C2 Aldehydes
Organism: Aeromonas sp. asnih1
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: SUGAR BINDING PROTEIN |
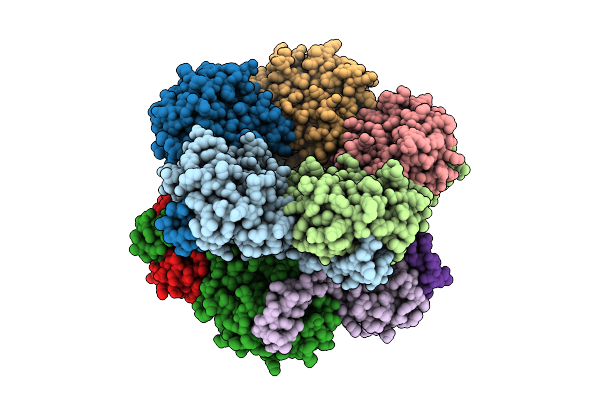 |
Selective Production Of Versatile L-Glyceraldehyde From C1 And/Or C2 Aldehydes
Organism: Gilliamella apicola
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: SUGAR BINDING PROTEIN |
 |
Human V-Atpase Vo Subcomplex (Containing Subunit Isoform A4) Bound To Nanobody And Inhibitor
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN Ligands: NAG, CLR, POV, A1A4Q, WJP |
 |
Organism: Homo sapiens, Human gammaherpesvirus 8, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: VIRAL PROTEIN/SIGNALING PROTEIN |
 |
Organism: Human gammaherpesvirus 8
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: ADP, ATP |
 |
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSFERASE |
 |
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: ACO, PO4 |
 |
Organism: Lama glama, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: PROTON TRANSPORT |
 |
Organism: Lama glama, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: PROTON TRANSPORT |
 |
Organism: Lama glama, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Resolution:3.70 Å Release Date: 2025-02-05 Classification: PROTON TRANSPORT |
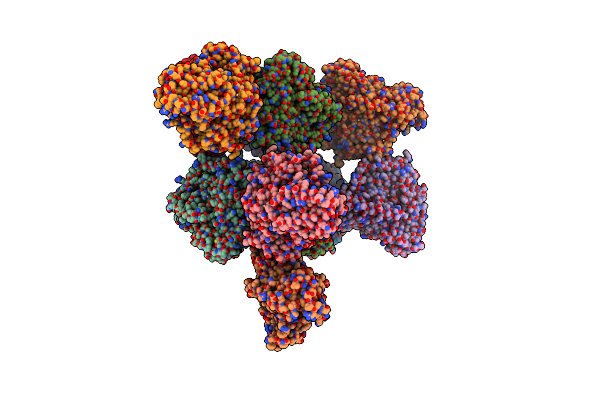 |
Organism: Enhygromyxa salina
Method: X-RAY DIFFRACTION Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: CD |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-11-22 Classification: LIGASE Ligands: FAD, TPP, MG, UQ0 |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2023-11-22 Classification: LIGASE Ligands: FAD, TPP, MG, UQ0 |
 |
Crystal Structure Of Escherichia Coli Glyoxylate Carboligase Double Mutant In Complex With Glycolaldehyde
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2023-11-22 Classification: LIGASE Ligands: MG, FAD, TPP, DW3, UQ1 |
 |
Crystal Structure Of Escherichia Coli Glyoxylate Carboligase Quadruple Mutant
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2023-11-22 Classification: LIGASE Ligands: FAD, TPP, MG, UQ0 |
 |
Organism: Sciscionella marina
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2023-02-22 Classification: HYDROLASE |
 |
Organism: Deinococcus metallilatus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-08-25 Classification: LIGASE |
 |
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-08-19 Classification: OXIDOREDUCTASE |
 |
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2020-08-19 Classification: OXIDOREDUCTASE |

