Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Aspartyl/Asparaginyl Beta-Hydroxylase (Asph) In Complex With Fe, 2-Oxoglutarate, Succinate And The Hydroxylated Product Of Factor X Derived Peptide Fragment, 12 H O2 Exposure
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: SIN, FE |
 |
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE |
 |
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE |
 |
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: PLP |
 |
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE |
 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens, Plasmodium falciparum
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: IMMUNE SYSTEM/DE NOVO PROTEIN |
 |
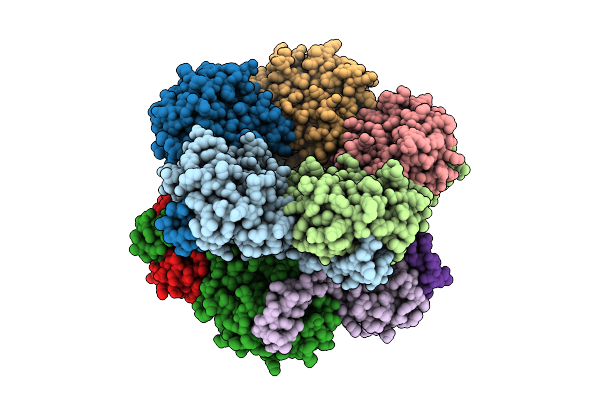
Selective Production Of Versatile L-Glyceraldehyde From C1 And/Or C2 Aldehydes
Organism: Aeromonas sp. asnih1
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: SUGAR BINDING PROTEIN |
 |
Selective Production Of Versatile L-Glyceraldehyde From C1 And/Or C2 Aldehydes
Organism: Gilliamella apicola
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: SUGAR BINDING PROTEIN |
 |
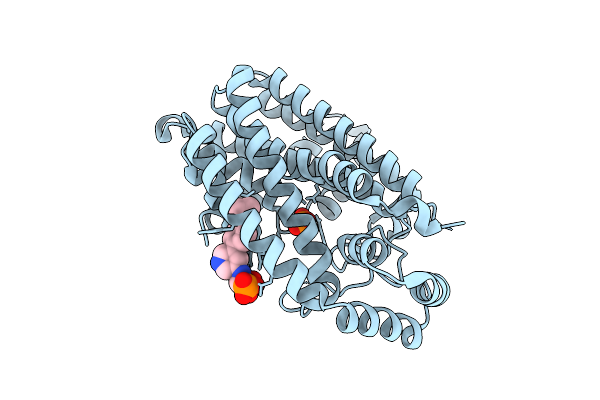
Serial Femtosecond Crystallography Structure Of Myoglobin From Equine Skeletal Muscle
Organism: Equus caballus
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXYGEN STORAGE Ligands: SO4, HEM |
 |
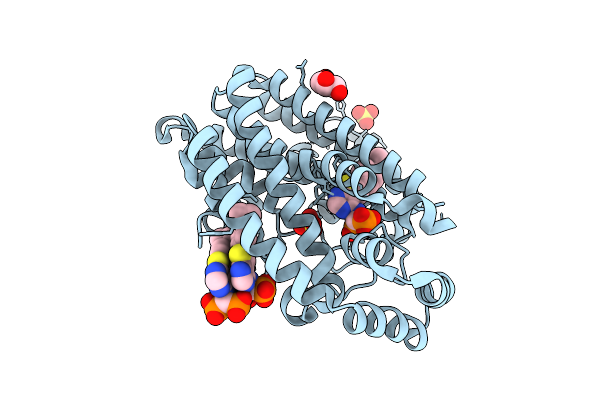
Crystal Structure Of Human Farnesyl Pyrophosphate Synthase In Complex With A Cryptic Pocket Ligand, Jds05120
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: PO4, JD5 |
 |
Crystal Structure Of Human Farnesyl Pyrophosphate Synthase In Complex With A Cryptic Pocket Ligand, Cl02134
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: GOL, SO4, YL2 |
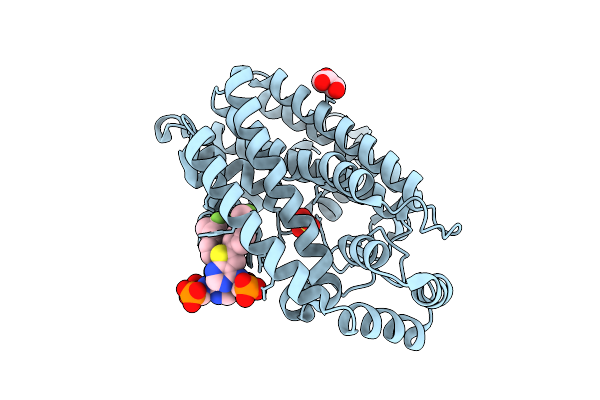 |
Crystal Structure (I) Of Human Farnesyl Pyrophosphate Synthase In Complex With A Cryptic Pocket Ligand, Cl03049
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: SO4, A1A05, GOL |
 |
Crystal Structure (Ii) Of Human Farnesyl Pyrophosphate Synthase In Complex With A Cryptic Pocket Ligand, Cl03049
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: A1A05, GOL, SO4 |
 |
Crystal Structure Of Au-15330 In Complex With The Bromodomain Of Human Brm (Smarca2) And Pvhl:Elonginc:Elonginb
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSCRIPTION Ligands: A1BNT, EDO, GOL |
 |
Crystal Structure Of Prt3789 In Complex With The Bromodomain Of Human Brg1 (Smarca4) And Pvhl:Elonginc:Elonginb
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSCRIPTION Ligands: A1BB4, GOL, EDO, CL |
 |
Crystal Structure Of Prt3789 In Complex With The Bromodomain Of Human Brm (Smarca2) And Pvhl:Elonginc:Elonginb
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSCRIPTION Ligands: A1BB4, CIT, ACT |
 |
Crystal Structure Of The Bromodomain Of Human Brm (Smarca2) In Complex With A Smarca-Bd Binder
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSCRIPTION Ligands: A1BB5, ZN |
 |
Organism: Clostridium phage phicd111
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: ANTIBIOTIC Ligands: ZN |