Search Count: 16
 |
Crystal Structure Of Mcl-1 In Complex With Computationally Designed Inhibitor Protein
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2022-07-20 Classification: APOTOSIS,DE NOVO PROTEIN |
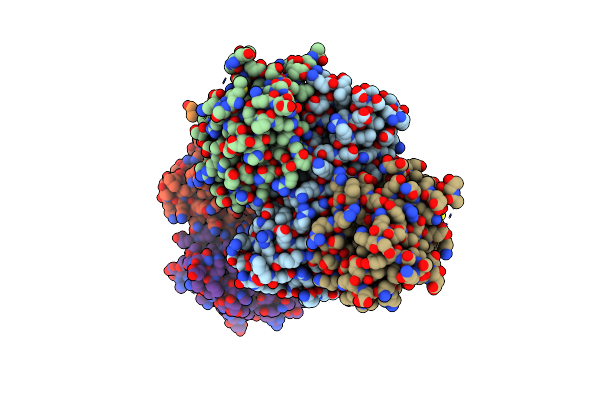 |
Crystal Structure Of Bcl-Xl In Complex With Computationally Designed Inhibitor Protein
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-07-20 Classification: APOTOSIS,DE NOVO PROTEIN |
 |
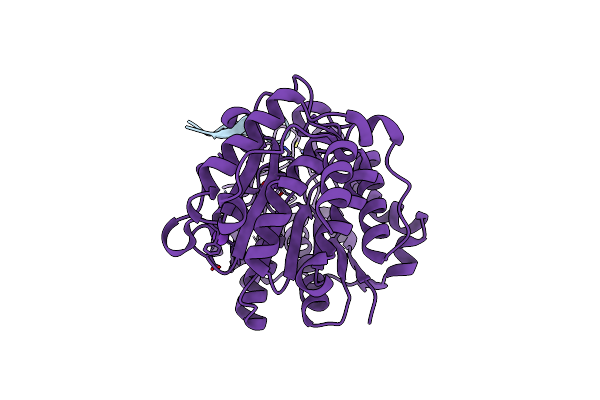
Crystal Structure Of Bcl-Xl In Complex With Computationally Designed Inhibitor Protein
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-07-20 Classification: APOTOSIS,DE NOVO PROTEIN |
 |
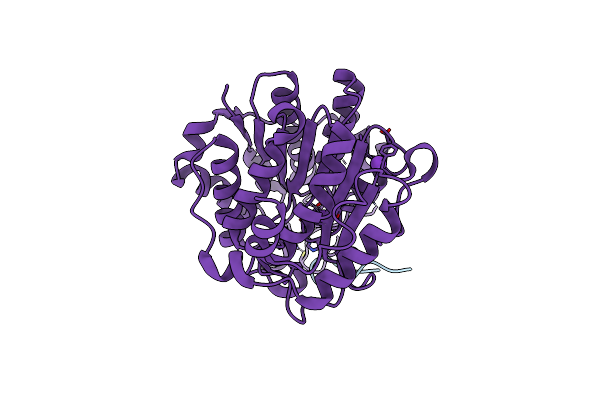
Crystal Structure Of Histone Deacetylase 4 (Hdac4) In Complex With A Smrt Corepressor Sp1 Fragment
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-11-14 Classification: HYDROLASE Ligands: K, ZN |
 |
Crystal Structure Of Histone Deacetylase 4 (Hdac4) In Complex With A Smrt Corepressor Sp2 Fragment
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-10-10 Classification: HYDROLASE Ligands: K, ZN |
 |
Crystal Structure Of Triosephosphate Isomerase From Thermoplasma Acidophilium
Organism: Thermoplasma acidophilum (strain atcc 25905 / dsm 1728 / jcm 9062 / nbrc 15155 / amrc-c165)
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2016-06-08 Classification: ISOMERASE Ligands: GOL, CL |
 |
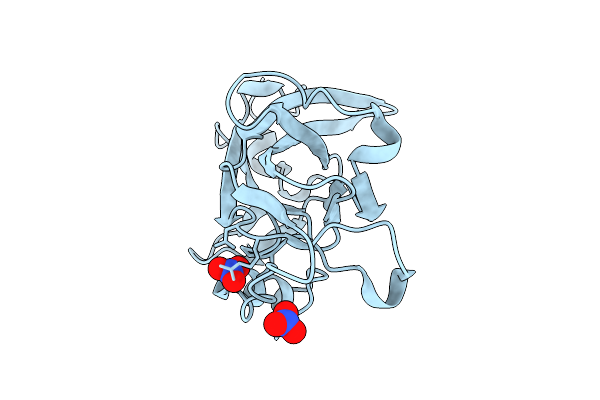
Crystal Structure Of Triosephosphate Isomerase From Thermoplasma Acidophilum With Glycerol 3-Phosphate
Organism: Thermoplasma acidophilum (strain atcc 25905 / dsm 1728 / jcm 9062 / nbrc 15155 / amrc-c165)
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2016-06-08 Classification: ISOMERASE Ligands: G3P, CL |
 |
Organism: Emericella nidulans
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2014-01-22 Classification: DNA BINDING PROTEIN Ligands: IOD, NO3 |
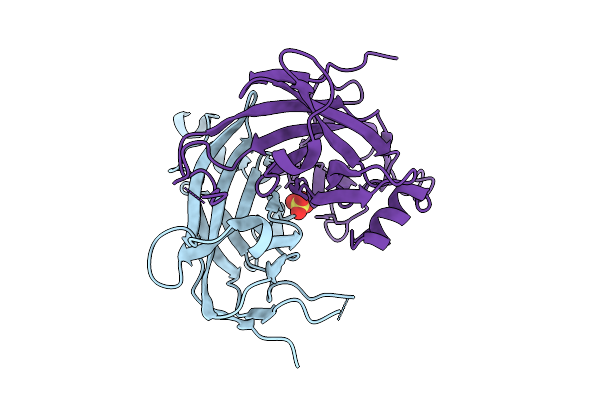 |
Organism: Emericella nidulans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-01-22 Classification: DNA BINDING PROTEIN/transcription Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-12-29 Classification: HYDROLASE |
 |
Organism: Antheraea pernyi
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2009-05-05 Classification: OXYGEN TRANSPORT Ligands: FMT |
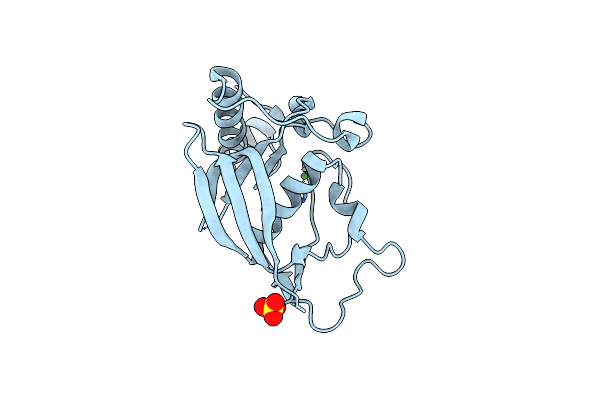 |
Structures Of Actinonin Bound Peptide Deformylases From E. Faecalis And S. Pyogenes
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2008-03-04 Classification: HYDROLASE Ligands: NI, SO4 |
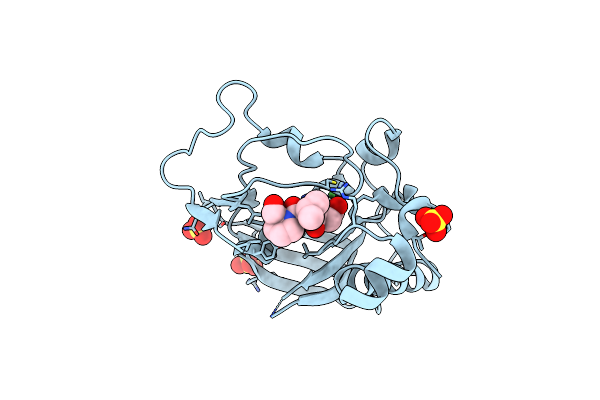 |
Structures Of Actinonin Bound Peptide Deformylases From E. Faecalis And S. Pyogenes
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2008-03-04 Classification: HYDROLASE Ligands: NI, SO4, BB2 |
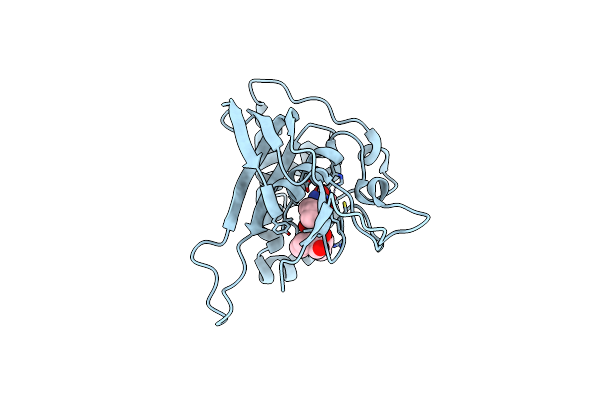 |
Structures Of Actinonin Bound Peptide Deformylases From E. Faecalis And S. Pyogenes
Organism: Streptococcus pyogenes m1 gas
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2008-03-04 Classification: HYDROLASE Ligands: CO, BB2 |
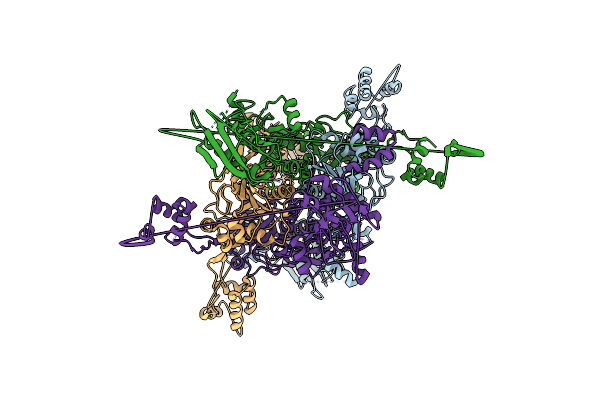 |
Organism: Methanococcus maripaludis
Method: X-RAY DIFFRACTION Resolution:3.23 Å Release Date: 2007-02-13 Classification: LIGASE |
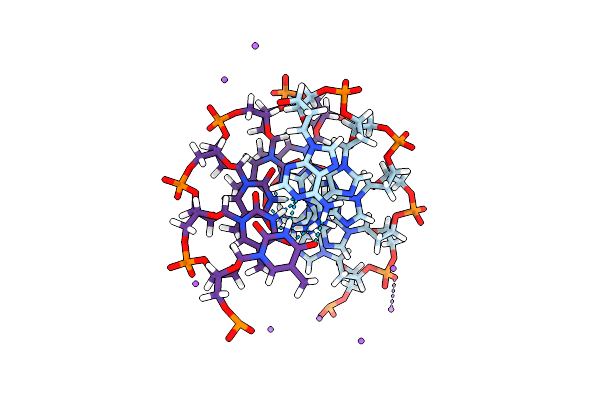 |
Sodium Ions And Water Molecules In The Structure Of Poly D(A)(Dot)Poly D(T)
|

