Search Count: 35
 |
Organism: Pseudomonas sp. mc1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: UNKNOWN FUNCTION |
 |
Monodehydroascorbate Reductase, Mdhar, From Antarctic Hairgrass Deschampsia Antarctica
Organism: Deschampsia antarctica
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-08-26 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Stenotrophomonas sp. kctc 12332
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2018-09-05 Classification: HYDROLASE |
 |
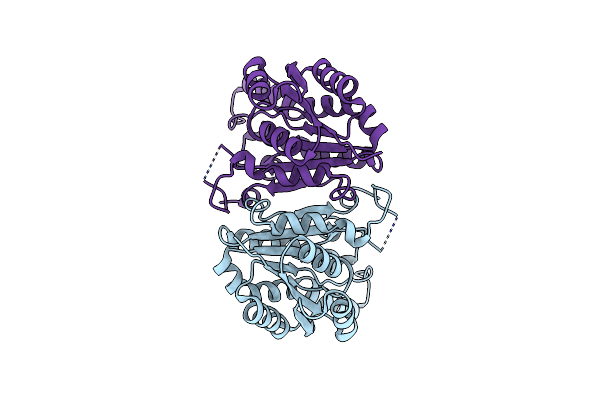
Crystal Structure Of Fabv, A New Class Of Enyl-Acyl Carrier Protein Reductase From Vibrio Fischeri
Organism: Vibrio fischeri (strain atcc 700601 / es114)
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2018-05-16 Classification: OXIDOREDUCTASE |
 |
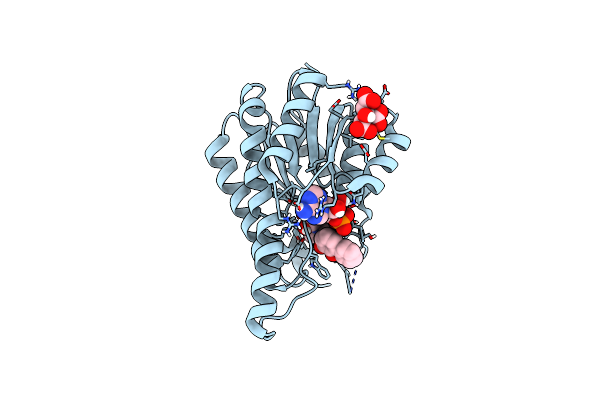
Crystal Structure Of The Cis-Dihydrodiol Naphthalene Dehydrogenase Nahb From Pseudomonas Sp. Mc1
Organism: Pseudomonas sp. mc1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-08-09 Classification: OXIDOREDUCTASE |
 |
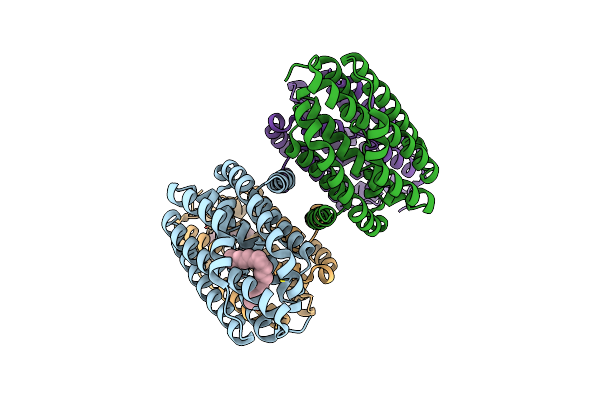
Crystal Structure Of The Cis-Dihydrodiol Naphthalene Dehydrogenase Nahb From Pseudomonas Sp. Mc1 In The Presence Of Nad+ And 2,3-Dihydroxybiphenyl
Organism: Pseudomonas sp. mc1
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2017-08-09 Classification: OXIDOREDUCTASE Ligands: NAD, CIT, BPY |
 |
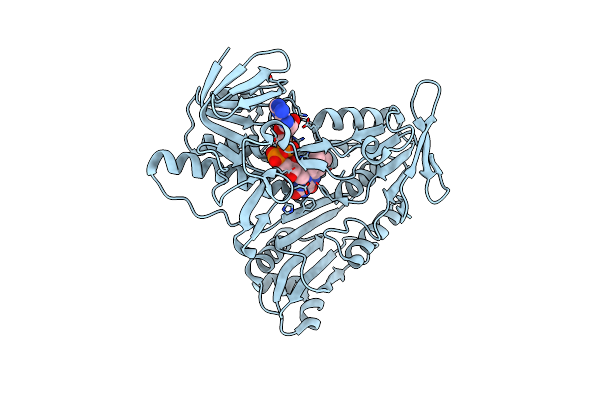
Crystal Structures Of Aldehyde Deformylating Oxygenase From Limnothrix Sp. Knua012
Organism: Limnothrix sp. knua012
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-03-15 Classification: METAL BINDING PROTEIN Ligands: OCD |
 |
Structure And Catalytic Mechanism Of Monodehydroascorbate Reductase, Mdhar, From Oryza Sativa L. Japonica
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-10-12 Classification: HYDROLASE Ligands: FAD |
 |
Structure And Catalytic Mechanism Of Monodehydroascorbate Reductase, Mdhar, From Oryza Sativa L. Japonica
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-10-12 Classification: HYDROLASE Ligands: FAD, NAD |
 |
Structure And Catalytic Mechanism Of Monodehydroascorbate Reductase, Mdhar, From Oryza Sativa L. Japonica
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-10-12 Classification: OXIDOREDUCTASE Ligands: FAD, NDP |
 |
Structure And Catalytic Mechanism Of Monodehydroascorbate Reductase, Mdhar, From Oryza Sativa L. Japonica
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-10-12 Classification: OXIDOREDUCTASE Ligands: FAD, NAD, ISD |
 |
Structure And Catalytic Mechanism Of Monodehydroascorbate Reductase, Mdhar, From Oryza Sativa L. Japonica
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2016-10-12 Classification: OXIDOREDUCTASE Ligands: FAD, NAD, ASC |
 |
Crystal Structures Of Aldehyde Deformylating Oxygenase From Oscillatoria Sp. Knua011
Organism: Oscillatoria sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-09-07 Classification: METAL BINDING PROTEIN Ligands: FE, STE |
 |
Structural Characterization Of The Full-Length Response Regulator Spr1814 In Complex With A Phosphate Analogue Reveals A Novel Conformational Plasticity Of The Linker Region
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-04-27 Classification: DNA BINDING PROTEIN Ligands: MG, BEF |
 |
Structure Of The Full-Length Response Regulator Spr1814 In Complex With A Phosphate Analogue And B3C
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-04-27 Classification: DNA BINDING PROTEIN Ligands: MG, BEF, 4QT |
 |
Crystal Structure Of Dehydroascorbate Reductase (Osdhar) From Oryza Sativa L. Japonica
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-02-03 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Oxidized Dehydroascorbate Reductase (Osdhar) From Oryza Sativa L. Japonica
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2016-02-03 Classification: PLANT PROTEIN Ligands: CA, PEG |
 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2016-02-03 Classification: PLANT PROTEIN Ligands: ASC |
 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2016-02-03 Classification: PLANT PROTEIN Ligands: GSH, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2015-08-12 Classification: TRANSFERASE |

