Search Count: 22
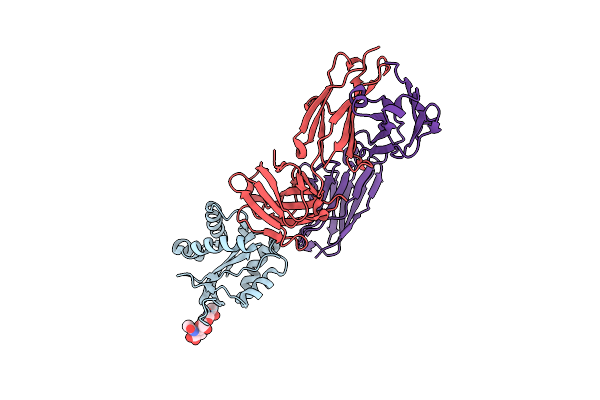 |
Crystal Structure Of Hiv-1 Mper Scaffold In Complex With Antibody Fab Ab45.1
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-02-19 Classification: VIRAL PROTEIN |
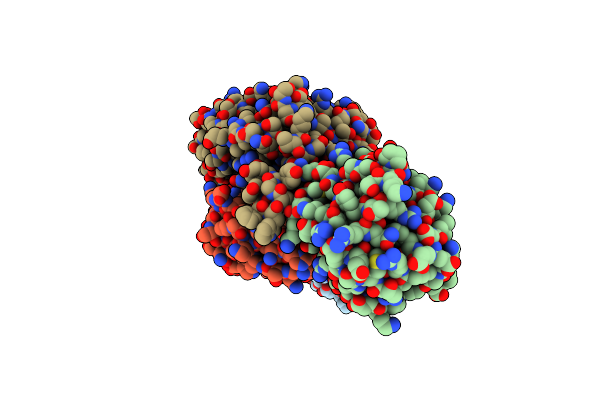 |
Crystal Structure Of Hiv-1 Mper Scaffold In Complex With Antibody Fab Ab45.2
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2025-02-19 Classification: VIRAL PROTEIN |
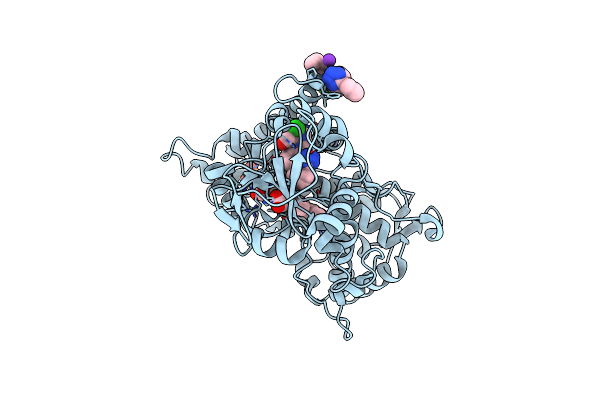 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2024-06-12 Classification: OXIDOREDUCTASE Ligands: LSN, K, HEM, PO4 |
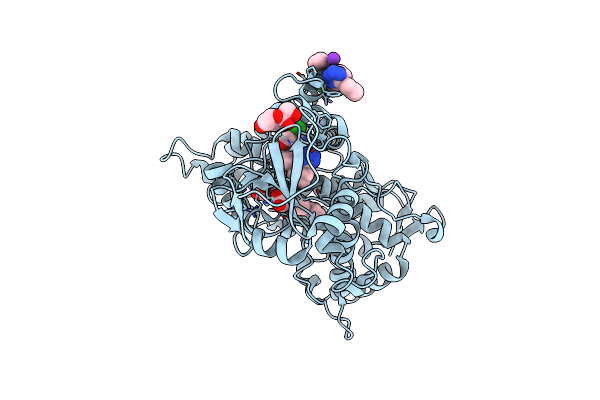 |
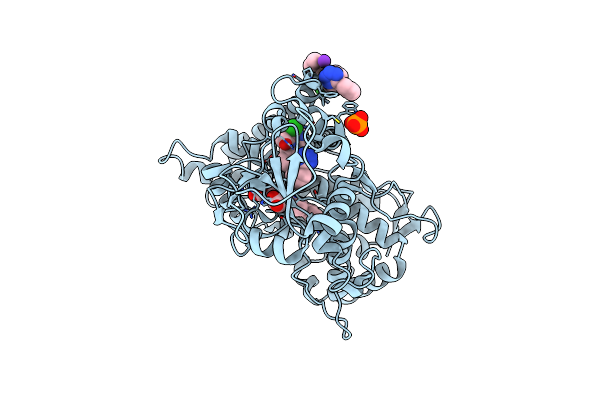
Crystal Structure Of Human Cytochrome P450 2C9*27 (R150L) Genetic Variant In Complex With The Drug Losartan
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2024-06-12 Classification: OXIDOREDUCTASE Ligands: HEM, LSN, FRU, K |
 |
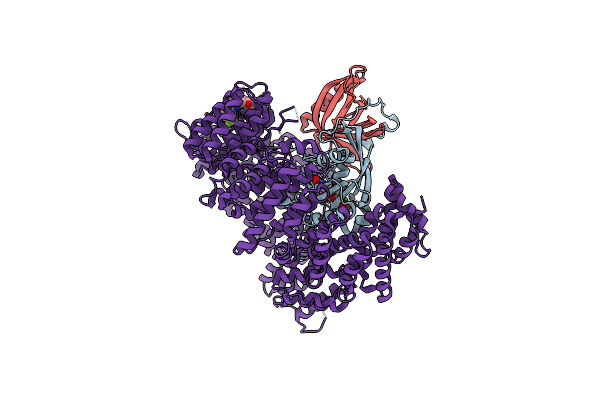
Crystal Structure Of The Human Cytochrome P450 2C9*8 (Cyp2C9*8) Genetic Variant In Complex With The Drug Losartan
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2021-10-06 Classification: OXIDOREDUCTASE Ligands: HEM, LSN, K, PO4 |
 |
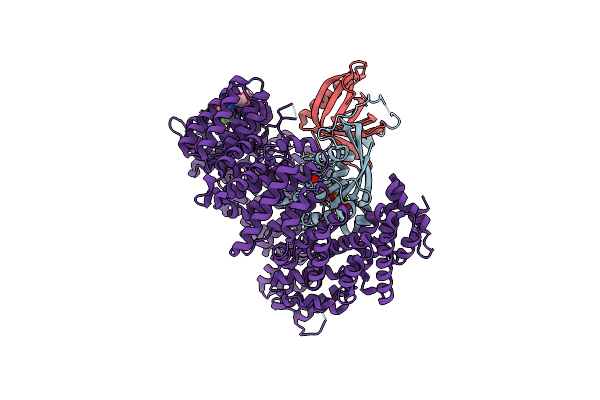
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-01-27 Classification: PROTEIN TRANSPORT Ligands: GNP, MG, K85 |
 |
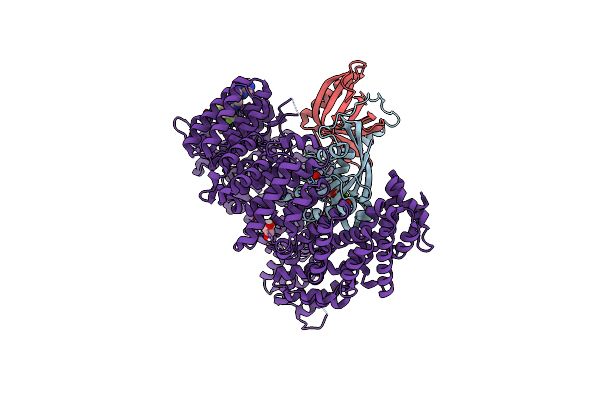
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-01-27 Classification: PROTEIN TRANSPORT Ligands: GNP, MG, K85 |
 |
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-01-27 Classification: PROTEIN TRANSPORT Ligands: GNP, MG, GOL, V6A |
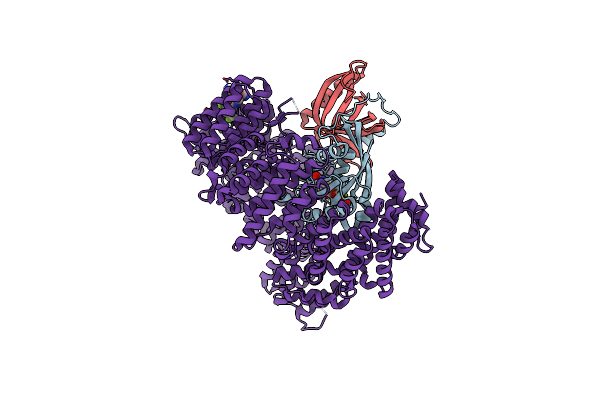 |
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2021-01-27 Classification: PROTEIN TRANSPORT Ligands: GNP, MG, 6L8 |
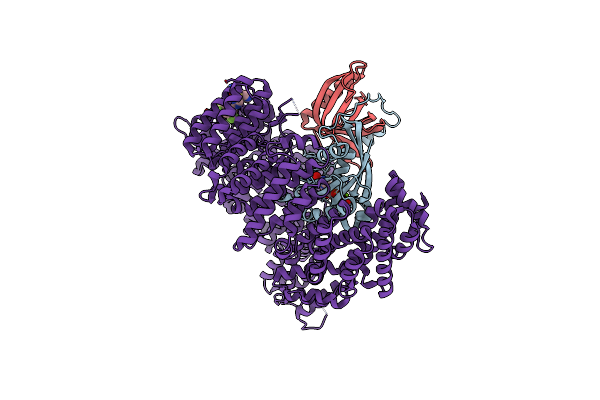 |
Crystal Structure Of Kpt-8602 Bound To Crm1 (E582K, 537-Dltvk-541 To Glceq)
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2021-01-27 Classification: PROTEIN TRANSPORT Ligands: GNP, MG, 6L8 |
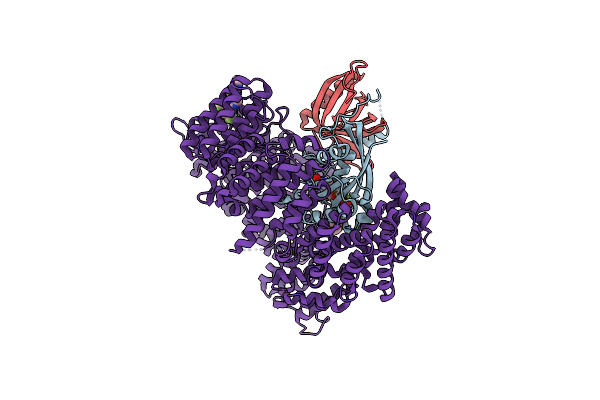 |
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-01-27 Classification: PROTEIN TRANSPORT Ligands: GNP, MG, GOL, V6A |
 |
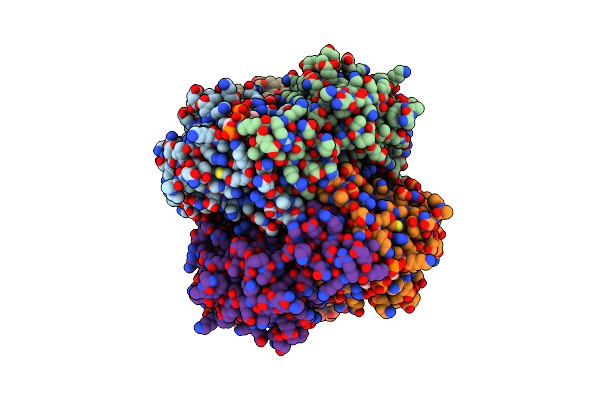
Crystal Structure Of Human P450 2C9*2 Genetic Variant In Complex With Losartan
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2020-09-30 Classification: OXIDOREDUCTASE Ligands: HEM, LSN, CM5, SO4 |
 |
Crystal Structure Analysis Of Rat Enoyl-Coa Hydratase In Complex With Hexadienoyl-Coa
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2002-09-24 Classification: LYASE Ligands: HXC |
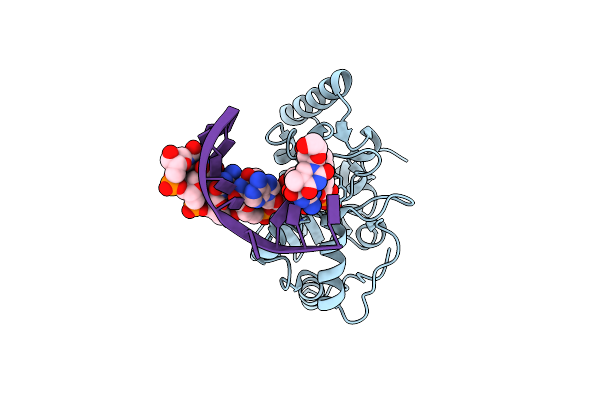 |
Crystal Structure Of Human Uracil-Dna Glycosylase Bound To Uncleaved Substrate-Containing Dna
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2000-05-16 Classification: hydrolase/DNA |
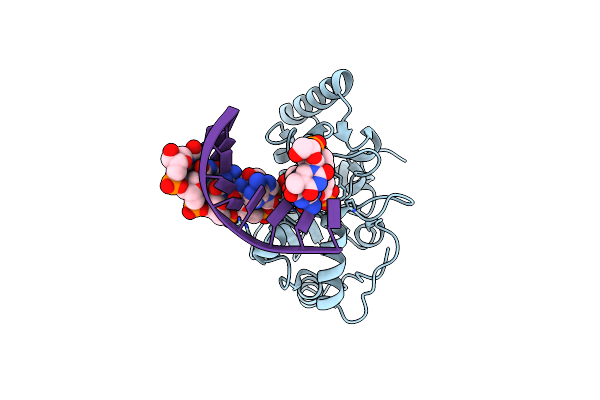 |
Uracil-Dna Glycosylase Bound To Dna Containing A 4'-Thio-2'Deoxyuridine Analog Product
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-05-16 Classification: hydrolase/DNA Ligands: URA |
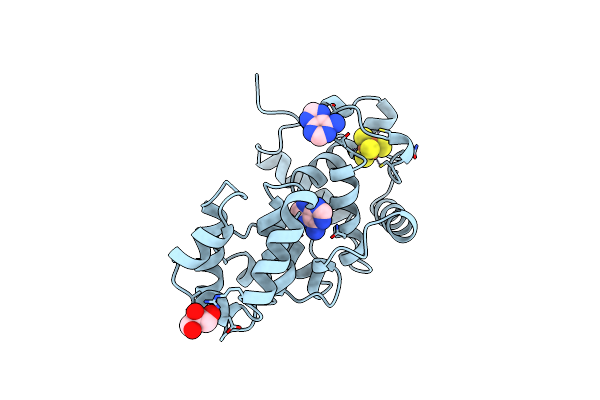 |
Catalytic Domain Of Muty From Escherichia Coli, D138N Mutant Complexed To Adenine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1999-09-20 Classification: HYDROLASE Ligands: SF4, ADE, GOL |
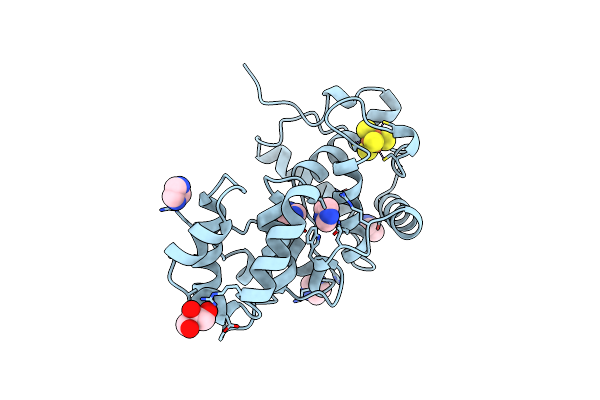 |
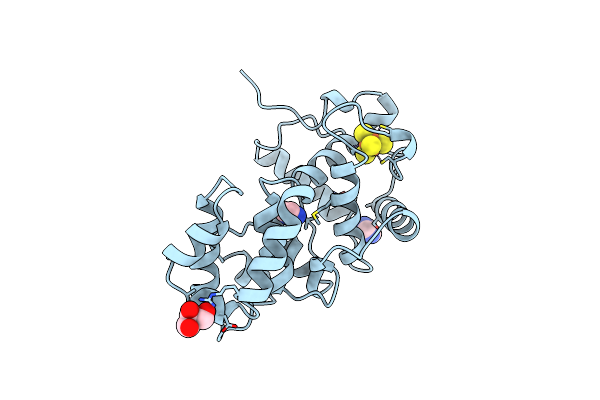
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 1999-08-26 Classification: DNA REPAIR Ligands: SF4, IMD, GOL |
 |
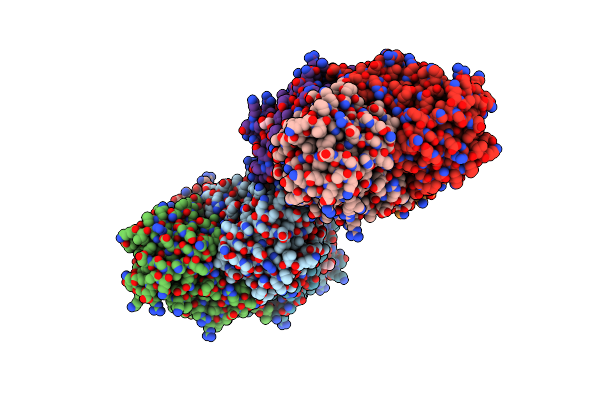
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 1999-08-20 Classification: DNA REPAIR Ligands: GOL, SF4, IMD |
 |
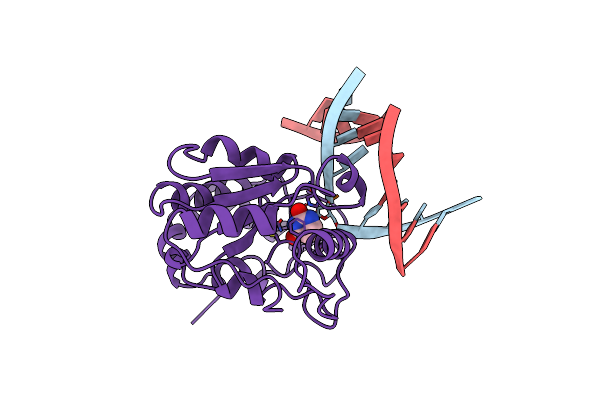
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 1999-07-21 Classification: OXIDOREDUCTASE Ligands: GLC, NAD, TCL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1999-05-06 Classification: HYDROLASE/DNA Ligands: URA |

