Search Count: 15
 |
Crystal Structure Of The Rna-Dependent Rna Polymerase 3D From Human Rhinovirus Serotype 14
Organism: Human rhinovirus 14
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2004-10-26 Classification: TRANSFERASE Ligands: SM |
 |
Crystal Structure Of Rna-Dependent Rna Polymerase 3D From Human Rhinovirus Serotype 1B
Organism: Human rhinovirus 1b
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2004-10-26 Classification: TRANSFERASE Ligands: K |
 |
Crystal Structure Of Rna-Dependent Rna Polymerase 3D From Human Rhinovirus Serotype 16
Organism: Human rhinovirus 16
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-10-26 Classification: TRANSFERASE |
 |
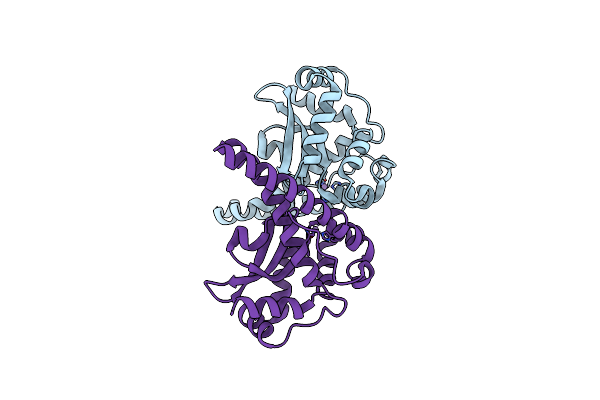
Crystal Structure Of Hcv Ns5B Rna Polymerase Complexed With A Novel Non-Competitive Inhibitor.
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-03-18 Classification: TRANSFERASE Ligands: NH1 |
 |
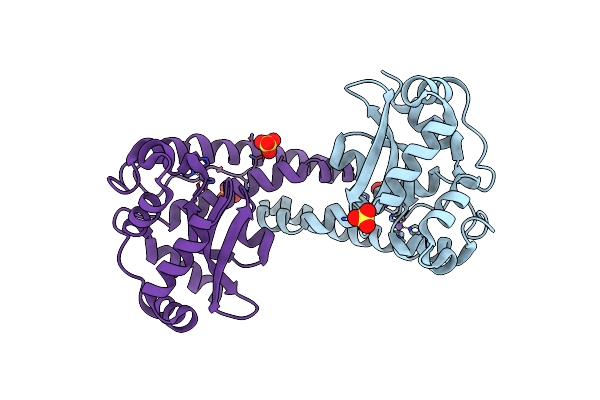
The Structure Of Human Mitochondrial Mn3+ Superoxide Dismutase Reveals A Novel Tetrameric Interface Of Two 4-Helix Bundles
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2002-11-06 Classification: OXIDOREDUCTASE Ligands: MN |
 |
Catalytic And Structural Effects Of Amino-Acid Substitution At His30 In Human Manganese Superoxide Dismutase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2002-11-06 Classification: OXIDOREDUCTASE Ligands: MN, SO4 |
 |
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 1998-04-29 Classification: CELL ADHESION Ligands: PT, HTO |
 |
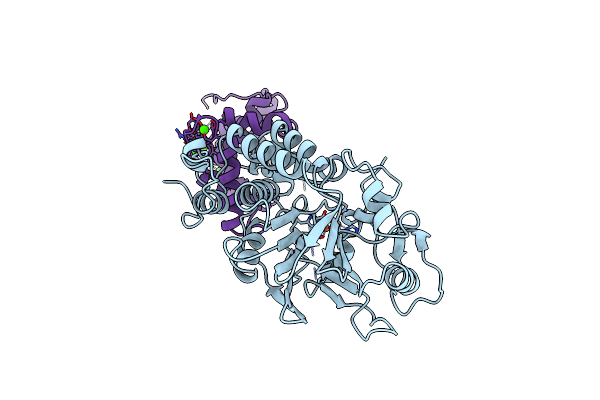
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1998-03-25 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-12-03 Classification: HYDROLASE Ligands: ZN, FE, CA |
 |
Mitochondrial Manganese Superoxide Dismutase Variant With Ile 58 Replaced By Thr
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1996-06-10 Classification: OXIDOREDUCTASE Ligands: MN3 |
 |
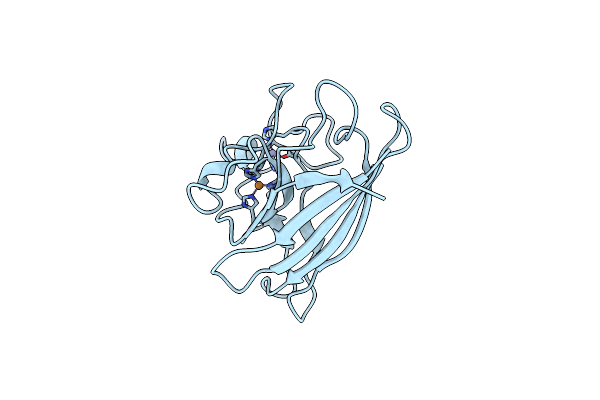
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 1996-06-10 Classification: OXIDOREDUCTASE (SUPEROXIDE ACCEPTOR) Ligands: CU1, ZN |
 |
Reduced Bridge-Broken Yeast Cu/Zn Superoxide Dismutase Low Temperature (-180C) Structure
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 1996-03-08 Classification: OXIDOREDUCTASE (SUPEROXIDE ACCEPTOR) Ligands: CU, ZN |
 |
Human Ckshs2 Atomic Structure: A Role For Its Hexameric Assembly In Cell Cycle Control
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1995-02-07 Classification: CELL DIVISION Ligands: SO4 |
 |
Amyotrophic Lateral Sclerosis And Structural Defects In Cu,Zn Superoxide Dismutase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1994-04-30 Classification: OXIDOREDUCTASE Ligands: CU, ZN |
 |
Atomic Structures Of Wild-Type And Thermostable Mutant Recombinant Human Cu, Zn Superoxide Dismutase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1993-04-15 Classification: OXIDOREDUCTASE (SUPEROXIDE ACCEPTOR) Ligands: CU, ZN, SO4 |

