Search Count: 31
 |
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-08-21 Classification: TRANSFERASE Ligands: GOL, CA, MG, O5R, EDO, PEG, BCN |
 |
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-09-06 Classification: TRANSFERASE Ligands: BCN, EDO, UDX, CA, MG, GOL |
 |
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-08-30 Classification: TRANSFERASE Ligands: GOL, EDO, MG, CA, BCN |
 |
Organism: Paramecium bursaria chlorella virus 1
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: VIRAL PROTEIN |
 |
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-07-06 Classification: BIOSYNTHETIC PROTEIN Ligands: DCM, DCP, ZN, MG |
 |
Chlorovirus Pbcv-1 Bi-Functional Dcmp/Dctp Deaminase Bi-Dcd With Dttp/Dtmp Bound
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-07-06 Classification: BIOSYNTHETIC PROTEIN Ligands: TTP, TMP, ZN, MG |
 |
Organism: Paramecium bursaria chlorella virus 1
Method: ELECTRON MICROSCOPY Release Date: 2019-01-30 Classification: VIRUS |
 |
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-10-18 Classification: VIRAL PROTEIN Ligands: HG, BGC |
 |
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2017-10-18 Classification: VIRAL PROTEIN Ligands: HG |
 |
The Crystal Structure Of Viral Collagen Prolyl Hydroxylase Vcph From Paramecium Bursaria Chlorella Virus-1 - 2Og Complex
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-09-30 Classification: OXIDOREDUCTASE Ligands: AKG, ZN, MN |
 |
The Crystal Structure Of Viral Collagen Prolyl Hydroxylase Vcph From Paramecium Bursaria Chlorella Virus-1 - Truncated Construct
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-09-30 Classification: HYDROLASE Ligands: MN, UNX, ACT |
 |
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2013-05-22 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: FAD, 0VJ, DMS |
 |
Paramecium Chlorella Bursaria Virus1 Putative Orf A654L Is A Polyamine Acetyltransferase
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-01-25 Classification: TRANSFERASE Ligands: COA, IMD |
 |
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-11-10 Classification: VIRAL PROTEIN Ligands: GOL, SO4, CL |
 |
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2010-11-10 Classification: VIRAL PROTEIN Ligands: SO4 |
 |
Organism: Paramecium bursaria chlorella virus 1, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2010-11-10 Classification: VIRAL PROTEIN Ligands: SAH |
 |
Crystal Structure Of A Glycosyltransferase Involved In The Glycosylation Of The Major Capsid Of Pbcv-1
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2007-08-21 Classification: TRANSFERASE Ligands: MN, FLC |
 |
Crystal Structure Of A Glycosyltransferase Involved In The Glycosylation Of The Major Capsid Of Pbcv-1
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-08-21 Classification: TRANSFERASE Ligands: MN, UPG |
 |
Crystal Structure Of A Glycosyltransferase Involved In The Glycosylation Of The Major Capsid Of Pbcv-1
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-08-21 Classification: TRANSFERASE Ligands: MN, UDP |
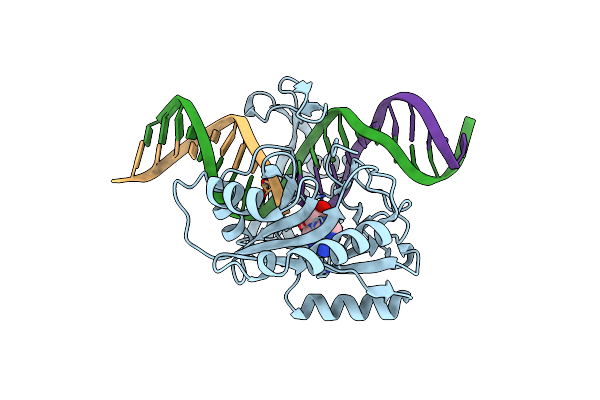 |
Structure Of Chlorella Virus Dna Ligase-Adenylate Bound To A 5' Phosphorylated Nick
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-07-10 Classification: LIGASE/DNA Ligands: AMP |

