Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Dokdonia eikasta
Method: ELECTRON MICROSCOPY Resolution:2.32 Å Release Date: 2025-01-29 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, RET, NA, OLA |
 |
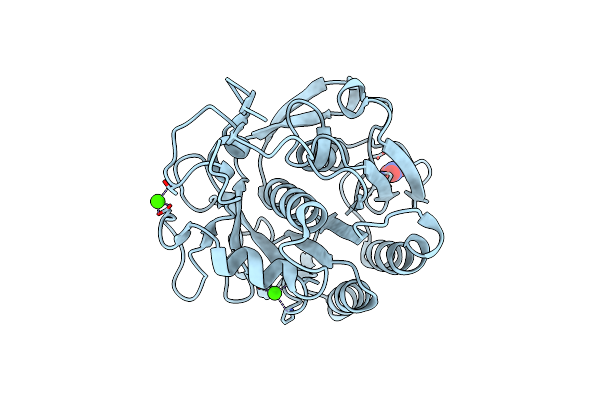
Serial Microseconds Crystallography At Id29 Using Sacla Extruder: Thaumatin
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-01-22 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-01-22 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-01-22 Classification: PLANT PROTEIN Ligands: TLA |
 |
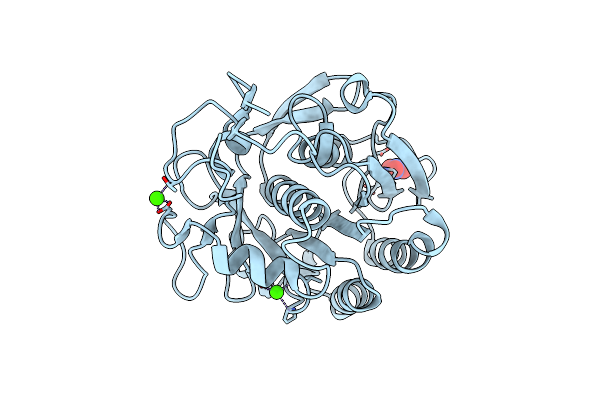
Serial Microseconds Crystallography At Id29 Using Fixed-Target (Small Foils): Proteinase K With 10 Um Spacing Between X-Ray Pulses
Organism: Parengyodontium album
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: CA, NO3 |
 |
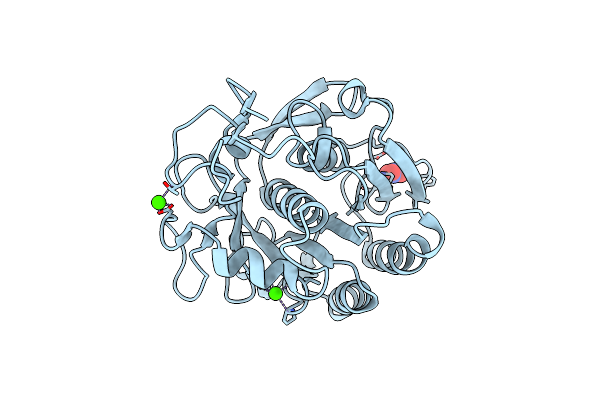
Serial Microseconds Crystallography At Id29 Using Fixed-Target (Small Foils): Proteinase K With 20 Um Spacing Between X-Ray Pulses
Organism: Parengyodontium album
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: CA, NO3 |
 |
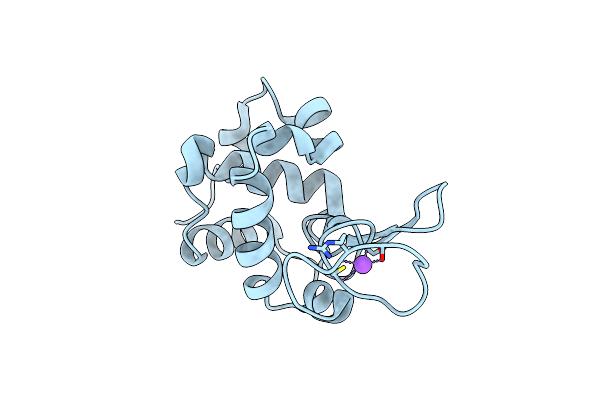
Serial Microseconds Crystallography At Id29 Using Fixed-Target (Large Foils): Proteinase K With 50 Um Spacing Between X-Ray Pulses
Organism: Parengyodontium album
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: CA, NO3 |
 |
Serial Microseconds Crystallography At Id29 Using Fixed-Target (Si Chip): Lysozyme - Without Ligand Glcnac (Apo)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2025-01-22 Classification: HYDROLASE |
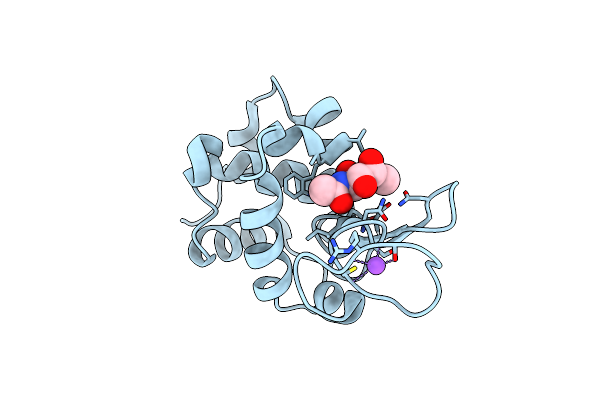 |
Serial Microseconds Crystallography At Id29 Using Fixed-Target (Si Chip): Lysozyme - With Ligand Glcnac
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2025-01-22 Classification: HYDROLASE |
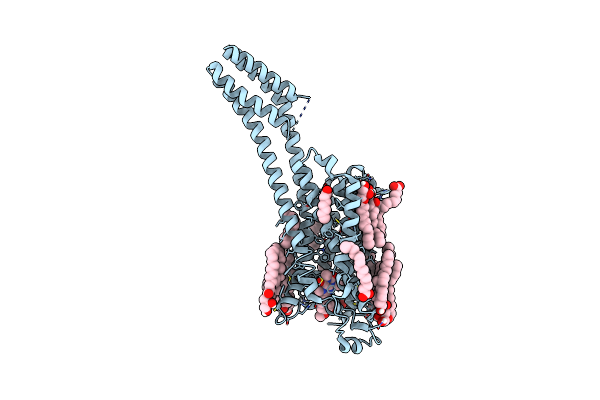 |
Serial Microseconds Crystallography At Id29 Using Fixed-Target (Small Foils): A2A Adenosine Receptor Co-Crystallised With Istradefylline
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-01-22 Classification: SIGNALING PROTEIN |
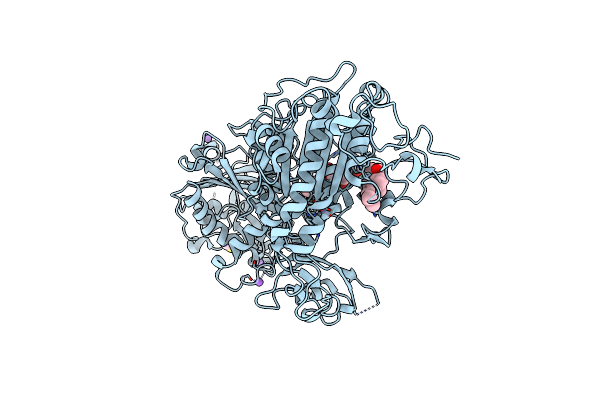 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: 5JK, SCN, ZN, CA, IOD, NA |
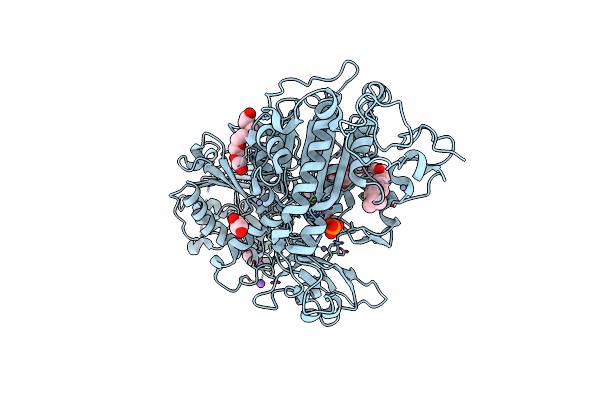 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: 5JK, GOL, 1PE, ZN, IOD, NA, CA, PO4 |
 |
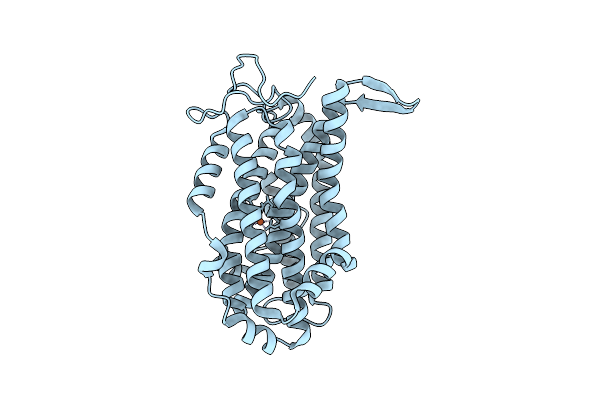
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-06-01 Classification: HORMONE |
 |
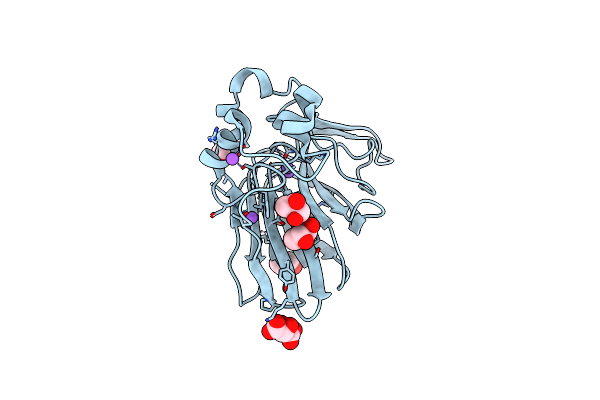
Structure Of Ribonucleotide Reductase R2 From Escherichia Coli Collected By Femtosecond Serial Crystallography On A Coc Membrane
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: FE |
 |
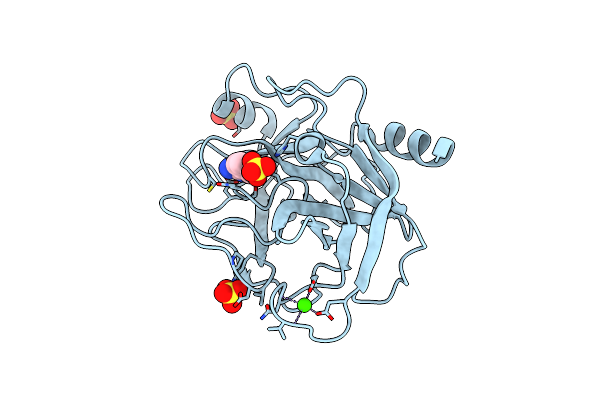
Structure Of Thaumatin Collected By Femtosecond Serial Crystallography On A Coc Membrane
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2021-11-24 Classification: PLANT PROTEIN Ligands: TLA, PGR, NA |
 |
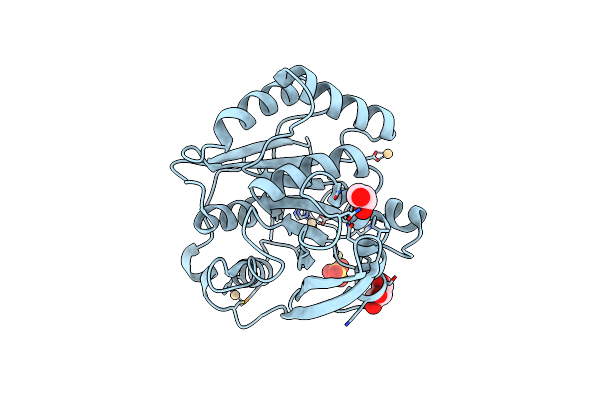
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-02-07 Classification: HYDROLASE Ligands: CA, BEN, SO4 |
 |
Structure Of Fae Solved By Sad From Data Collected At The Peak Of The Selenium Absorption Edge On Id30B
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-02-07 Classification: HYDROLASE Ligands: CD, GOL, SO4 |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2018-02-07 Classification: PLANT PROTEIN Ligands: TLA, GOL |
 |
Structure Of Thermolysin Solved From Sad Data Collected At The Peak Of The Zn Absorption Edge On Id30B
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2018-01-31 Classification: HYDROLASE Ligands: ZN, CA, DMS, TMO, VAL, LYS |
 |
Structure Of Thaumatin Collected From An In Situ Crystal Collected On Id30B At 12.7 Kev.
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-01-31 Classification: PLANT PROTEIN Ligands: TLA |