Search Count: 15
 |
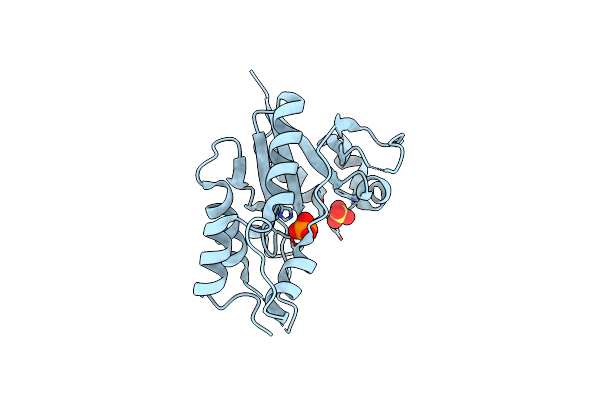
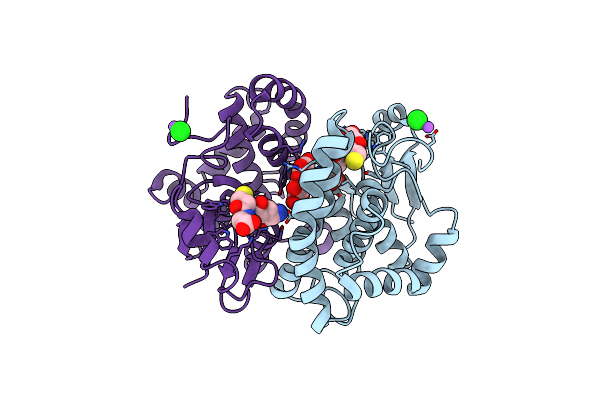
Structural Insights Into The Nmn Complex Of Nicotinate Nucleotide Adenylyltransferase From Enterococcus Faecium Via Co-Crystallization Studies
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2024-07-17 Classification: TRANSFERASE Ligands: NMN, PGE, PEG, PO4, MG |
 |
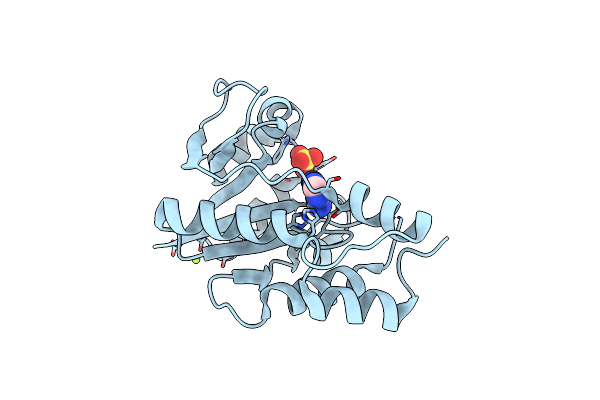
Crystal Structure Of Enterococcus Faecium Nicotinate Nucleotide Adenylyltransferase At 1.9 Angstroms Resolution
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-08-16 Classification: TRANSFERASE Ligands: SO4, 2HP |
 |
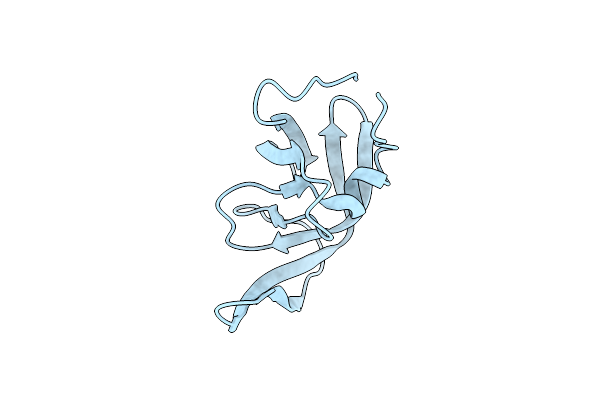
High Resolution Crystal Structure Of Enterococcus Faecium Nicotinate Nucleotide Adenylyltransferase Complexed With Adenine
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2023-08-16 Classification: TRANSFERASE Ligands: SO4, ADE, MG |
 |
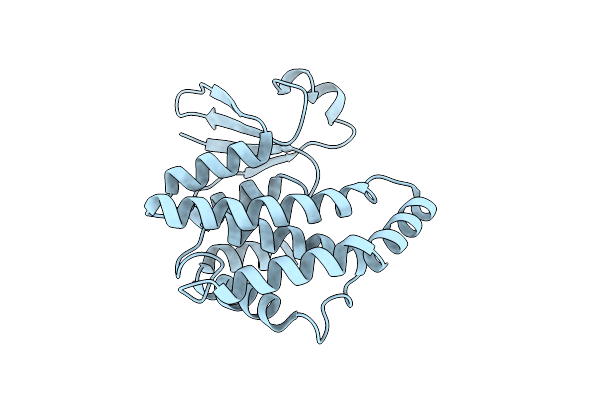
Apo-Crystal Structure Of A Wild-Type South African Hiv-1 Subtype C Protease At 2.4 Angstrom
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-04-12 Classification: HYDROLASE |
 |
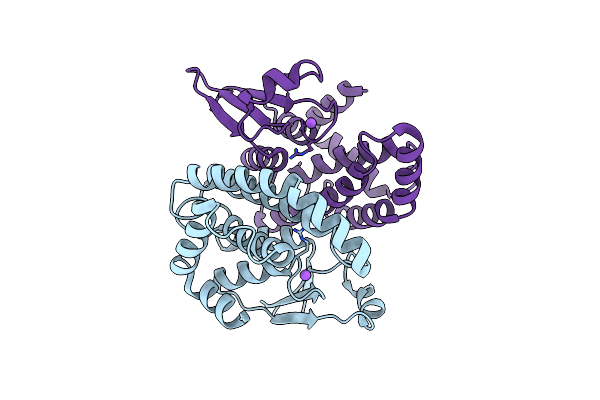
The Apo-Crystal Structure Of A Variant Form Of The 28-Kda Schistosoma Bovis Glutathione Transferase In Orthorhombic Form
Organism: Schistosoma bovis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-11-30 Classification: TRANSFERASE |
 |
The Apo-Crystal Structure Of A Variant Form Of The 28-Kda Schistosoma Haematobium Glutathione Transferase
Organism: Schistosoma haematobium
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-11-02 Classification: TRANSFERASE |
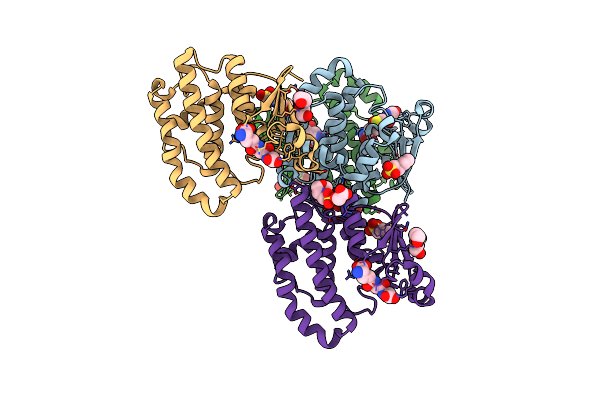 |
Crystal Structure Of Human Glutathione Transferase P1-1 (Hgstp1-1) That Was Co-Crystallised In The Presence Of Indanyloxyacetic Acid-94 (Iaa-94)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-03-11 Classification: TRANSFERASE Ligands: GSH, MES, O7Z, PGE |
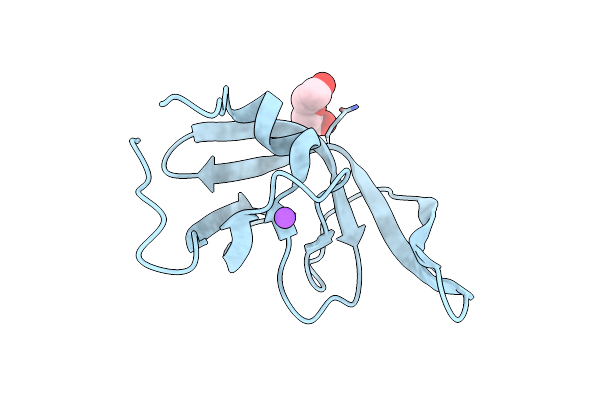 |
Crystal Structure Of I13V/I62V/V77I South African Hiv-1 Subtype C Protease Containing A D25A Mutation
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-02-26 Classification: HYDROLASE |
 |
Crystal Structure Of Sjgst In Complex With Gsh And Ellagic Acid At 1.53 Angstrom Resolution
Organism: Schistosoma japonicum
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2019-06-19 Classification: TRANSFERASE Ligands: GSH, NA, CL, REF |
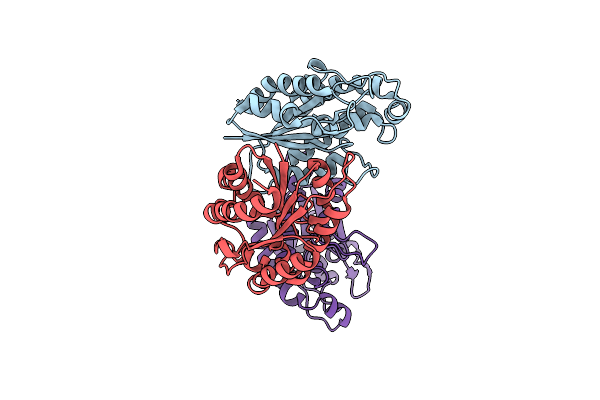 |
The Crystal Structure Of Carbohydrate Acetylesterase Family Member From Sinorhizobium Meliloti
Organism: Sinorhizobium meliloti 1021
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-12-24 Classification: HYDROLASE |
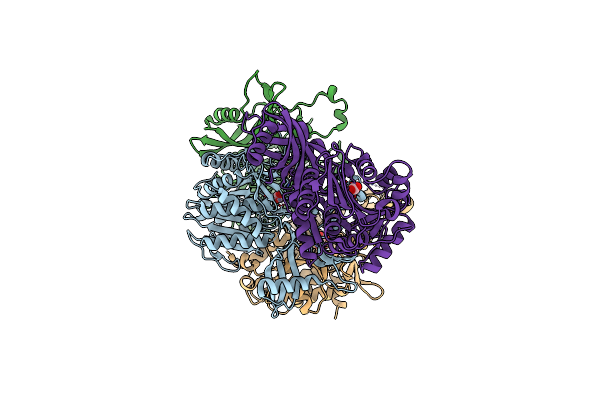 |
Structural And Kinetic Bases For The Metal Preference Of The M18 Aminopeptidase From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-04-16 Classification: HYDROLASE Ligands: ZN, CO3 |
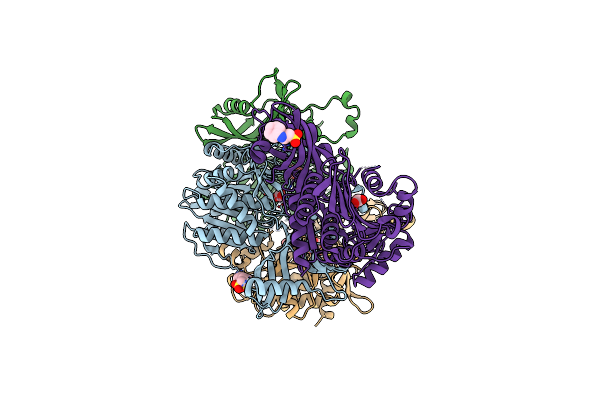 |
Structural And Kinetic Bases For The Metal Preference Of The M18 Aminopeptidase From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-04-02 Classification: HYDROLASE Ligands: CO, CO3, NHE |
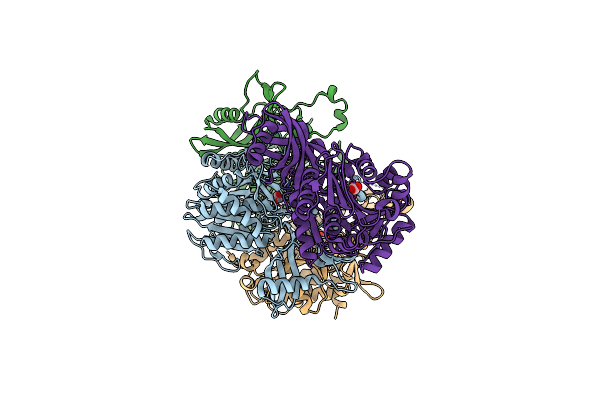 |
Structural And Kinetic Bases For The Metal Preference Of The M18 Aminopeptidase From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-04-02 Classification: HYDROLASE Ligands: ZN, CO3 |
 |
Structural And Kinetic Bases For The Metal Preference Of The M18 Aminopeptidase From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-04-02 Classification: HYDROLASE |
 |
Structural And Kinetic Bases For The Metal Preference Of The M18 Aminopeptidase From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2014-04-02 Classification: HYDROLASE Ligands: ZN |

