Search Count: 133
 |
Organism: Dissulfurispira thermophila
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: IMMUNE SYSTEM Ligands: ZN |
 |
Organism: Escherichia coli, Salmonella enterica
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: IMMUNE SYSTEM |
 |
Organism: Salmonella enterica
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: IMMUNE SYSTEM |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-03-26 Classification: TRANSFERASE Ligands: MG, ADP |
 |
Crystal Structure Of Guanine Nucleotide-Binding Protein (G Protein) Alpha-1 Subunit From Selaginella Moellendorffii In Complex With Gtp Gamma S
Organism: Selaginella moellendorffii
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-11-06 Classification: SIGNALING PROTEIN Ligands: MG, GSP |
 |
Crystal Structure Of Guanine Nucleotide-Binding Protein Alpha Subunit (G Protein) From Oryza Sativa In Complex With Gdp
Organism: Oryza sativa indica group
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2024-11-06 Classification: SIGNALING PROTEIN Ligands: GDP, MG |
 |
Hmpv Fusion Protein Complexed With Single Domain Antibodies Sdhmpv16 And Sdhmpv12
Organism: Lama glama, Human metapneumovirus a
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-01-24 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
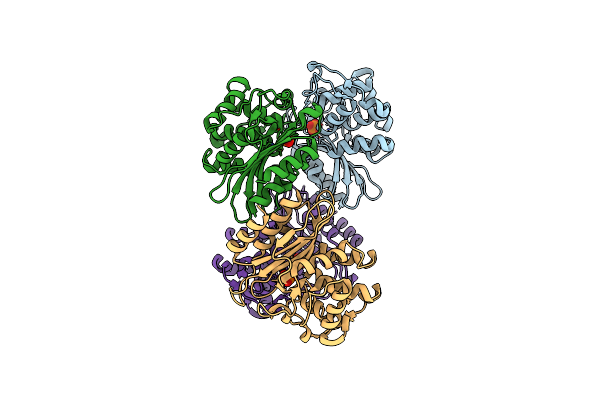
Xfel Structure Of Mycobacterium Tuberculosis Beta Lactamase Microcrystals Mixed With Sulbactam For 3 Ms
Organism: Mycobacterium tuberculosis str. beijing/w bt1
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2023-09-20 Classification: HYDROLASE/Inhibitor Ligands: PO4 |
 |
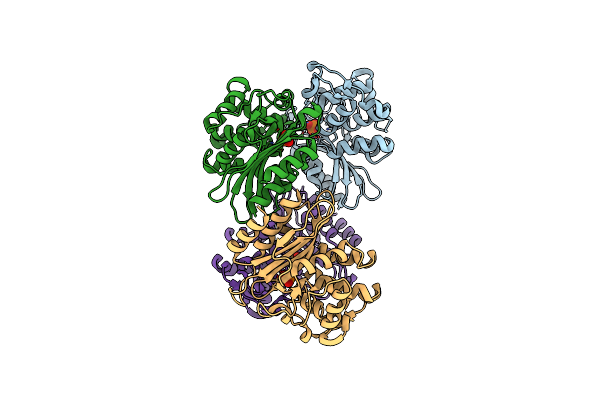
Xfel Structure Of Mycobacterium Tuberculosis Beta Lactamase Microcrystals Mixed With Sulbactam For 6 Ms
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2023-09-20 Classification: HYDROLASE/Inhibitor Ligands: PO4 |
 |
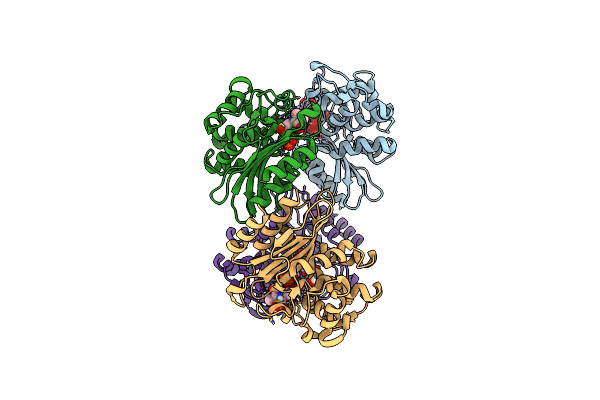
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-09-20 Classification: HYDROLASE/Inhibitor Ligands: PO4 |
 |
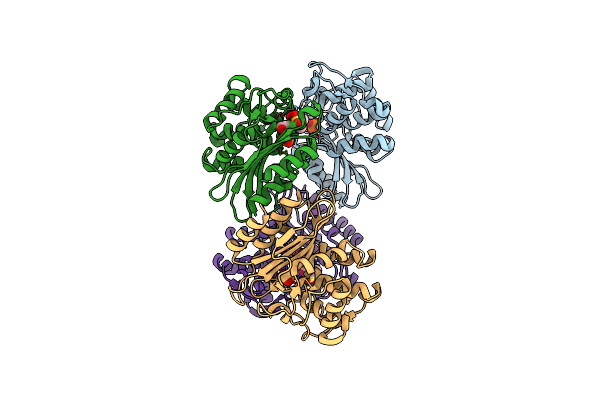
Xfel Structure Of Mycobacterium Tuberculosis Beta Lactamase Microcrystals Mixed With Sulbactam For 700 Ms
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2023-09-20 Classification: HYDROLASE/Inhibitor Ligands: PO4, TSL |
 |
Xfel Structure Of Beta Lactamase Microcrystals Mixed With Sulbactam Solution For 15Ms
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-03-08 Classification: HYDROLASE Ligands: PO4, TSL |
 |
Xfel Structure Of Mycobacterium Tuberculosis Beta Lactamase Microcrystals Mixed With Sulbactam For 30Ms
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-03-08 Classification: HYDROLASE/Inhibitor Ligands: PO4, 0RN, TSL |
 |
Xfel Structure Of Mycobacterium Tuberculosis Beta Lactamase Microcrystals Mixed With Sulbactam For 240Ms
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-03-08 Classification: HYDROLASE/Inhibitor Ligands: PO4, TSL |
 |
Cryo Structure Of Mycobacterium Tuberculosis Beta Lactamase Microcrystals Mixed With Sulbactam For 3 Hours
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2023-03-08 Classification: HYDROLASE/Inhibitor Ligands: PO4, TSL |
 |
Crystal Structure Of Multi-Functional Polysaccharide Lyase Smlt1473 From Stenotrophomonas Maltophilia (Strain K279A) In Apo Form At Ph 8.5
Organism: Stenotrophomonas maltophilia k279a
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2021-10-27 Classification: LYASE Ligands: SO4 |
 |
Crystal Structure Of Multi-Functional Polysaccharide Lyase Smlt1473 (Wt) From Stenotrophomonas Maltophilia (Strain K279A) In Apo Form At Ph 6.5
Organism: Stenotrophomonas maltophilia k279a
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2021-10-27 Classification: LYASE Ligands: SO4, PEG |
 |
Crystal Structure Of Multi-Functional Polysaccharide Lyase Smlt1473 From Stenotrophomonas Maltophilia (Strain K279A) In Apo Form At Ph 5.5
Organism: Stenotrophomonas maltophilia k279a
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2021-10-27 Classification: LYASE |
 |
Crystal Structure Of Multi-Functional Polysaccharide Lyase Smlt1473 (Wt) From Stenotrophomonas Maltophilia (Strain K279A) In Apo Form At Ph 5.0
Organism: Stenotrophomonas maltophilia k279a
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2021-10-27 Classification: LYASE Ligands: SO4, PEG |
 |
Crystal Structure Of Multi-Functional Polysaccharide Lyase Smlt1473 (Wt) From Stenotrophomonas Maltophilia (Strain K279A) In Apo Form At Ph 7.0
Organism: Stenotrophomonas maltophilia k279a
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-10-27 Classification: LYASE Ligands: SO4 |

