Search Count: 28
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-07-30 Classification: BIOSYNTHETIC PROTEIN Ligands: FES |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-07-30 Classification: BIOSYNTHETIC PROTEIN Ligands: ALA, ZN |
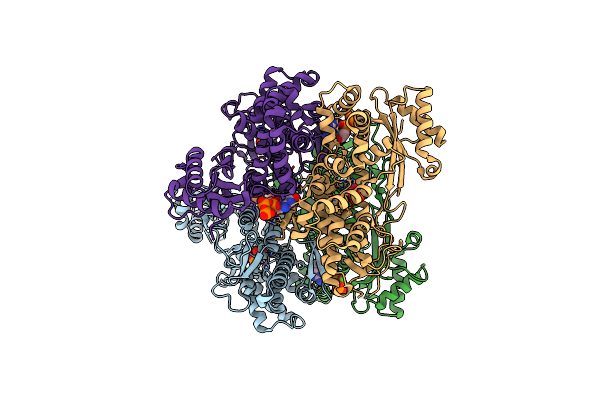 |
Organism: Rhizophagus irregularis
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2025-04-30 Classification: ANTIVIRAL PROTEIN Ligands: GTP, POP, MN, CA |
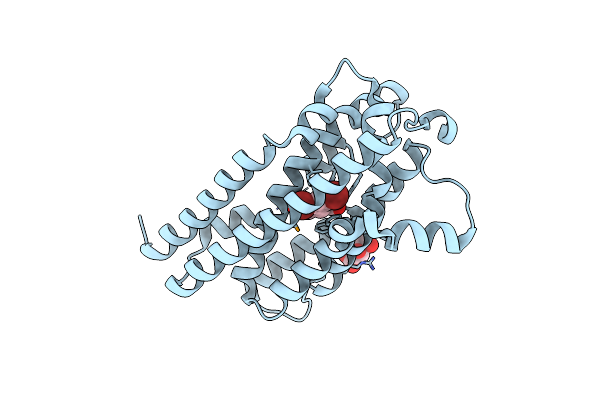 |
Organism: Aetokthonos hydrillicola
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2024-08-28 Classification: OXIDOREDUCTASE Ligands: 67I, MLT |
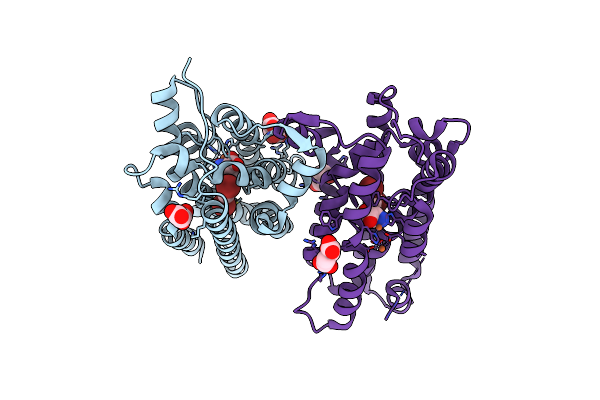 |
Crystal Structure Of Nitrile Synthase Aetd With Substrate Bound And Cofactor Partially Assembled
Organism: Aetokthonos hydrillicola
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-08-28 Classification: OXIDOREDUCTASE Ligands: 67I, MLT, SIN, FE, GOL |
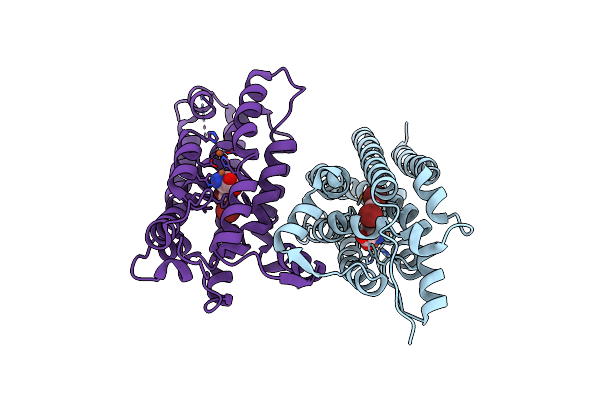 |
Crystal Structure Of Nitrile Synthase Aetd With Substrate Bound And Cofactor Fully Assembled
Organism: Aetokthonos hydrillicola
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-08-28 Classification: OXIDOREDUCTASE Ligands: 67I, FE |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-07-31 Classification: OXIDOREDUCTASE Ligands: FE2, K, MG, GOL |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2024-07-31 Classification: OXIDOREDUCTASE Ligands: FE2 |
 |
Crystal Structure Of Flcd From Pseudomonas Aeruginosa Bound To Iron (Ii) And Substrate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2024-07-31 Classification: OXIDOREDUCTASE Ligands: A1AL2, FE |
 |
Crystal Structure Of Flcd From Pseudomonas Aeruginosa Bond To Iron(Ii) And Substrate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-07-31 Classification: OXIDOREDUCTASE Ligands: FE2, A1AL2 |
 |
Organism: Flavobacterium johnsoniae (strain atcc 17061 / dsm 2064 / uw101)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-04-17 Classification: OXIDOREDUCTASE |
 |
Organism: Actinobacillus ureae atcc 25976
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2019-04-17 Classification: OXIDOREDUCTASE Ligands: CL, MG, EDO, GOL |
 |
X-Ray Crystal Structure Of Tmpb, (R)-1-Hydroxy-2-Trimethylaminoethylphosphonate Oxygenase, With (R)-1-Hydroxy-2-Trimethylaminoethylphosphonate
Organism: Leisingera caerulea
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2019-03-06 Classification: OXIDOREDUCTASE Ligands: FE, FE2, KVP |
 |
X-Ray Crystal Structure Of Tmpa, 2-Trimethylaminoethylphosphonate Hydroxylase, With Fe And 2Og
Organism: Leisingera caerulea
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2019-03-06 Classification: OXIDOREDUCTASE Ligands: FE2, AKG, SO4 |
 |
X-Ray Crystal Structure Of Tmpa, 2-Trimethylaminoethylphosphonate Hydroxylase, With Fe, 2Og, And 2-Trimethylaminoethylphosphonate
Organism: Leisingera caerulea
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-03-06 Classification: OXIDOREDUCTASE Ligands: FE2, AKG, KVV, SO4 |
 |
X-Ray Crystal Structure Of Tmpa, 2-Trimethylaminoethylphosphonate Hydroxylase, With Fe, 2Og, And (R)-1-Hydroxy-2-Trimethylaminoethylphosphonate
Organism: Leisingera caerulea
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2019-03-06 Classification: OXIDOREDUCTASE Ligands: FE2, KVP |
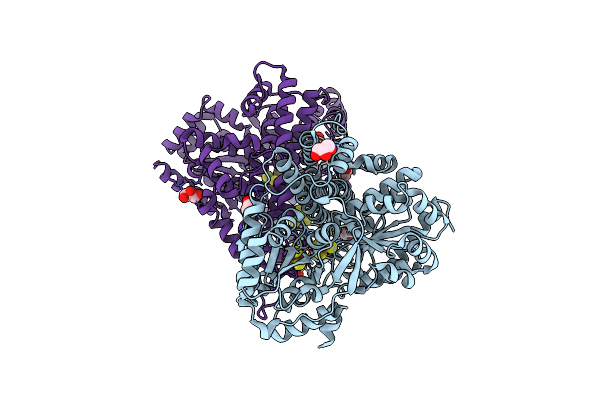 |
Crystal Structure Of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, As-Isolated (Protein Batch 1), Canonical C-Cluster
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-10-03 Classification: OXIDOREDUCTASE Ligands: SF4, XCC, GOL, FES |
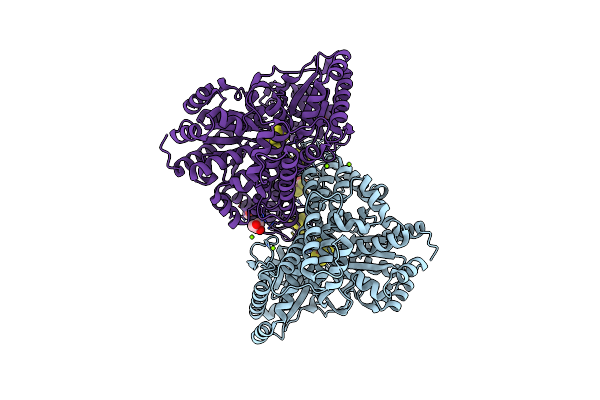 |
Crystal Structure Of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, As-Isolated (Protein Batch 2), Oxidized C-Cluster
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2018-10-03 Classification: OXIDOREDUCTASE Ligands: SF4, MG, XCC, CUV, FES, GOL, CL |
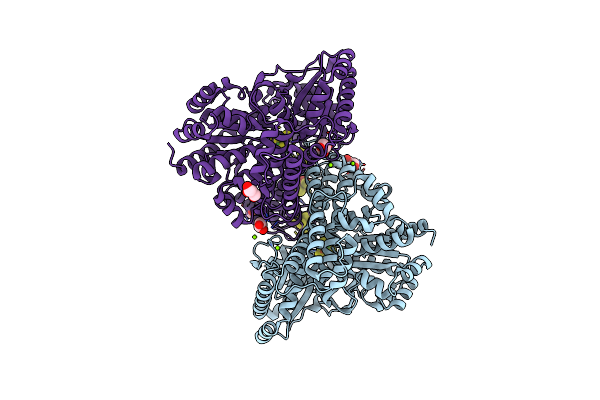 |
Crystal Structure Of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced (Protein Batch 2), Canonical C-Cluster
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2018-10-03 Classification: OXIDOREDUCTASE Ligands: SF4, XCC, GOL, MG, CL, FES |
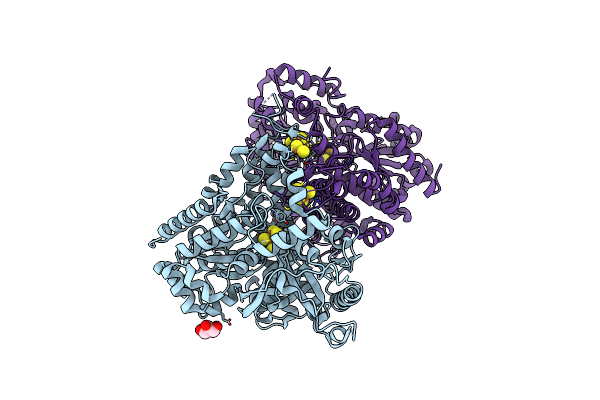 |
Crystal Structure Of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced Then Oxygen-Exposed (Protein Batch 2), Oxidized C-Cluster
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-10-03 Classification: OXIDOREDUCTASE Ligands: SF4, CUV, GOL, FES |

