Search Count: 617
 |
Organism: Escherichia coli o127:h6
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: P6G, PE8, GOL, PEG |
 |
Crystal Structure Of De Novo Designed Light-Responsive Oligomer C2-5 (Lro-C2-5)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of De Novo Designed Light-Responsive Oligomer C2-35 (Lro-C2-35) At Acidic Ph (Ph 4.5)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of De Novo Designed Light-Responsive Oligomer C2-35 (Lro-C2-35) At Basic Ph (Ph 8.5)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN Ligands: SO4 |
 |
Crystal Structure Of De Novo Designed Light-Responsive Oligomer C3-7 (Lro-C3-7)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of De Novo Designed Light-Responsive Oligomer C4-13 (Lro-C4-13)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of De Novo Designed Light-Responsive Oligomer C5-1 (Lro-C5-1)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of De Novo Designed Light-Responsive Heterodimer 2 (Lrd-2)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of De Novo Designed Light-Responsive Heterodimer 7 (Lrd-7)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: TRANSFERASE Ligands: NMN, GOL, PEG, EDO |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Atp|Adp-Bound Ifasym-2 State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ATP, MG, ADP |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Adp-Bound Ifasym-3 State (Atp 37 Degrees C Treated
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ADP, MG |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Adp-Bound Ifasym-3 State (Adp 4 Degrees C Treated)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ADP, MG |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Adp-Bound Ifasym-3 (Peptidisc) State (Atp 37Degrees C Treated)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ADP |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Adp-Bound Ifasym-3 (Peptidisc) State (Adp 4Degrees C Treated)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ADP |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Atp|Adp+Vi-Bound Occ (Vi) State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ATP, MG, ADP, VO4 |
 |
Cryo-Em Structure Of Msrv1273C/72C(E553Q) Mutant From Mycobacterium Smegmatis In The Atp-Bound Occ State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ATP, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: HYDROLASE |
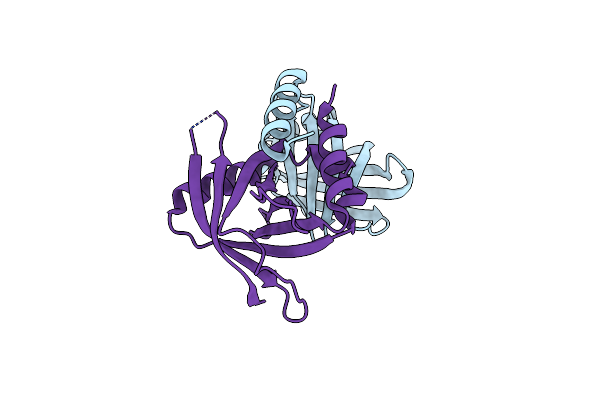 |
Crystal Structure Of Human Respiratory Syncytial Virus Ns1 Bound To Human Med25 Acid
Organism: Human respiratory syncytial virus a, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2025-04-09 Classification: VIRAL PROTEIN |
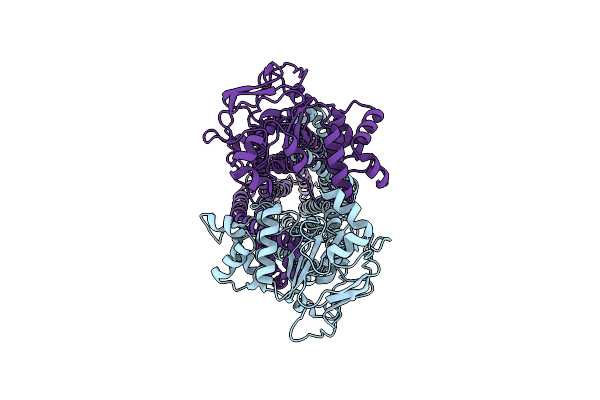 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Ifapo State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: TRANSPORT PROTEIN |

