Search Count: 17
 |
Organism: Gloeocapsa sp. pcc 7513
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2022-04-13 Classification: PHOTOSYNTHESIS Ligands: 45D |
 |
Organism: Gluconobacter oxydans
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-03-08 Classification: OXIDOREDUCTASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2015-07-15 Classification: OXIDOREDUCTASE Ligands: NI, AKG, GOL |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-07-15 Classification: TRANSCRIPTION Ligands: NI, AKG |
 |
Organism: Escherichia coli, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-04-30 Classification: LIPID BINDING PROTEIN Ligands: BOG, MG |
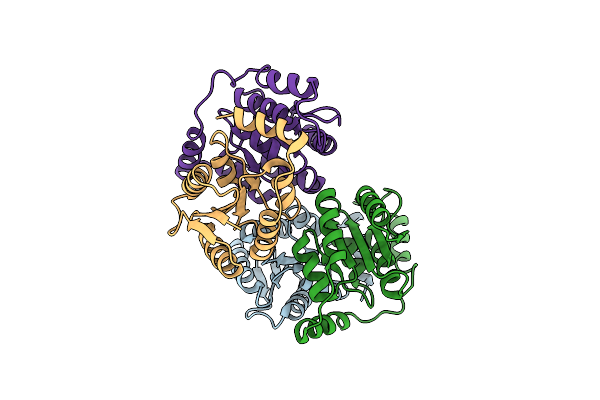 |
Crystal Structure Of D-Galactose-6-Phosphate Isomerase In A Substrate-Free Form
Organism: Lactobacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2013-11-27 Classification: ISOMERASE |
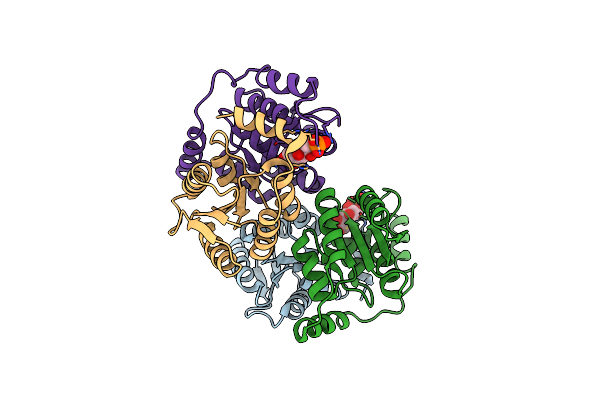 |
Crystal Structure Of D-Galactose-6-Phosphate Isomerase In Complex With D-Tagatose-6-Phosphate
Organism: Lactobacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2013-11-27 Classification: ISOMERASE Ligands: TG6 |
 |
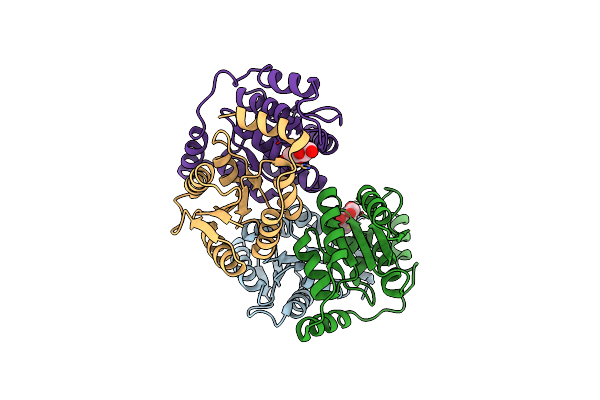
Crystal Structure Of D-Galactose-6-Phosphate Isomerase In Complex With D-Psicose
Organism: Lactobacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2013-11-27 Classification: ISOMERASE Ligands: PSJ |
 |
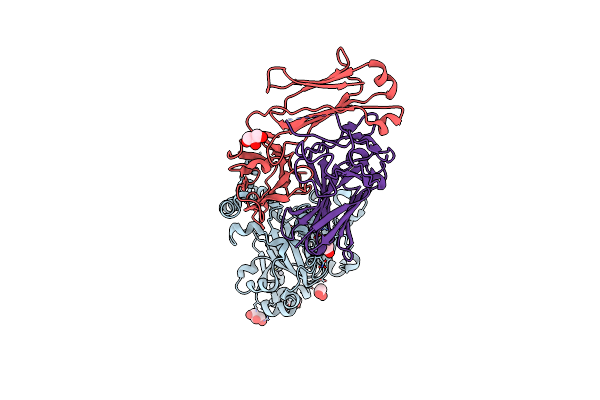
Crystal Structure Of D-Galactose-6-Phosphate Isomerase In Complex With D-Ribulose
Organism: Lactobacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2013-11-27 Classification: ISOMERASE Ligands: RBL |
 |
Organism: Homo sapiens, Cricetulus migratorius
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-08-04 Classification: IMMUNE SYSTEM Ligands: HG, GOL, SO4, FE |
 |
Organism: Homo sapiens, Cricetulus migratorius
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-08-04 Classification: IMMUNE SYSTEM Ligands: GOL, SO4, AKG, FE, HG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2006-05-23 Classification: METAL BINDING PROTEIN Ligands: ZN, FE2 |
 |
Crystal Structure Of Catalytic Core Domain Of Jmjd2A Complexed With Alpha-Ketoglutarate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2006-05-23 Classification: METAL BINDING PROTEIN Ligands: ZN, FE2, AKG |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2006-01-31 Classification: TRANSCRIPTION |
 |
Crystal Structure Of Sara, A Transcription Regulator From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2006-01-31 Classification: TRANSCRIPTION Ligands: CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2003-05-13 Classification: IMMUNE RESPONSE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2003-05-13 Classification: IMMUNE SYSTEM |

