Search Count: 29
 |
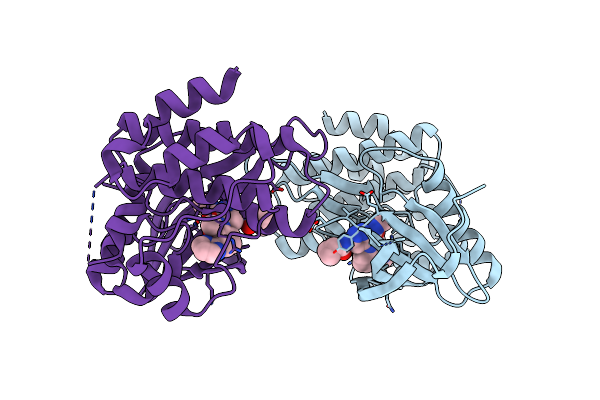
Organism: Rattus norvegicus, Ovis aries
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2021-12-01 Classification: CELL CYCLE Ligands: SO4, GTP, MG, GDP, N9B |
 |
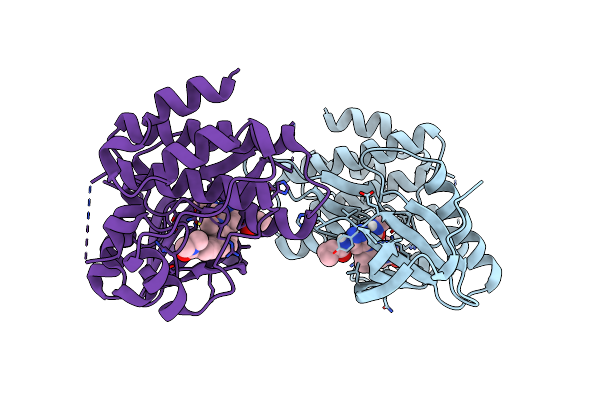
Crystal Structure Of Tyrosine Kinase 2 Jh2 (Pseudo Kinase Domain) Complexed With Compound-12 Aka:6-[(Cyclopropanecarbonyl)Amino]-4-{[2-Methoxy-3-(Pyrimidin-2-Yl)Phenyl]Amino}-N-Methylpyridazine-3-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2021-08-04 Classification: TRANSFERASE Ligands: VZJ |
 |
Crystal Structure Of Tyrosine Kinase 2 Jh2 (Pseudo Kinase Domain) Complexed With Compound-12 Aka:6-[(Cyclopropanecarbonyl)Amino]-4-({3-[6-(Dimethylcarbamoyl)Pyridazin-3-Yl]-2-Methoxyphenyl}Amino)-N-Methylpyridazine-3-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2021-08-04 Classification: TRANSFERASE Ligands: VZG |
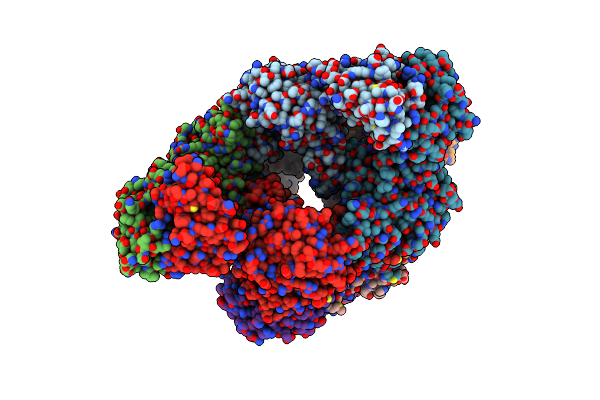 |
Organism: pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2019-02-27 Classification: OXIDOREDUCTASE |
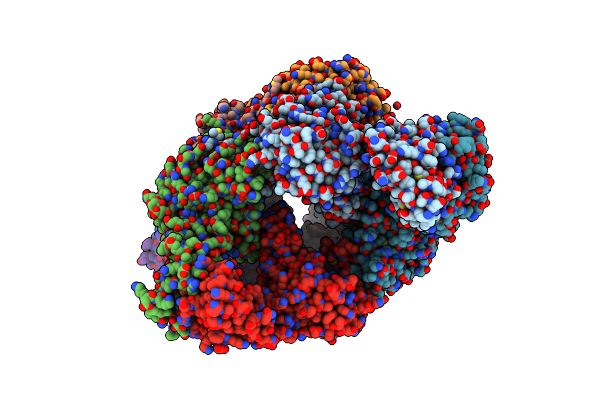 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2019-02-27 Classification: OXIDOREDUCTASE Ligands: F2K, SO4 |
 |
Organism: Enterococcus faecalis (strain atcc 700802 / v583)
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2017-12-27 Classification: OXIDOREDUCTASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-06-28 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Cpap Pn2-3 C-Terminal Loop-Helix In Complex With Darpin-Tubulin
Organism: Synthetic construct, Homo sapiens, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-10-26 Classification: CELL CYCLE Ligands: GTP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-09-14 Classification: UNKNOWN FUNCTION Ligands: GOL, IOD |
 |
Organism: Streptomyces lividans, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2012-04-18 Classification: MEMBRANE PROTEIN Ligands: K, CD |
 |
Organism: Streptomyces lividans, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-04-18 Classification: MEMBRANE PROTEIN Ligands: MTN, K |
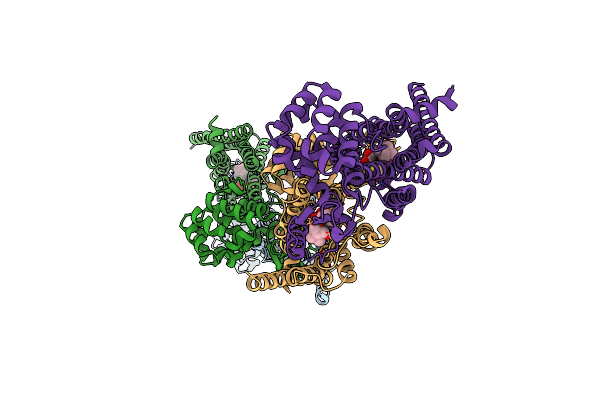 |
Organism: Rattus norvegicus, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2012-02-22 Classification: SIGNALING PROTEIN, Hydrolase Ligands: 0HK, PO4 |
 |
Hcv Ns3-4A Protease Domain With A Ketoamide Inhibitor Derivative Of Boceprevir Bound
Organism: Hepatitis c virus subtype 1a
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2011-02-23 Classification: Hydrolase/Hydrolase Inhibitor Ligands: ZN, MCX, BME |
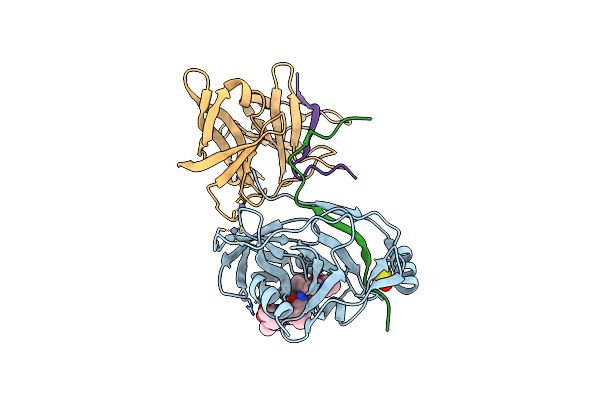 |
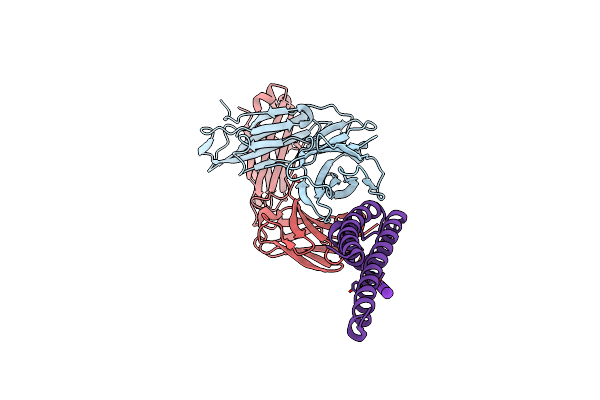
Organism: Hepatitis c virus subtype 1a
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2011-01-19 Classification: VIRAL PROTEIN/INHIBITOR Ligands: ZN, NNA, BME |
 |
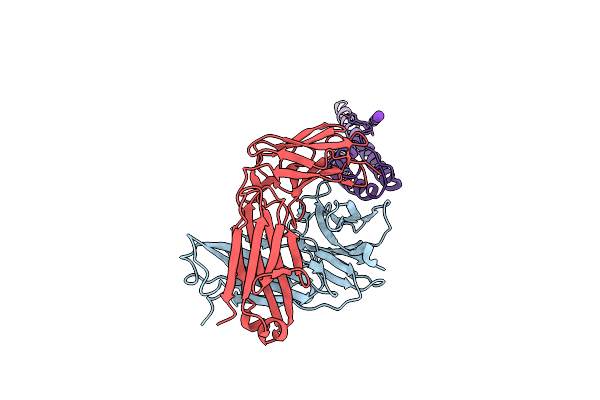
Organism: Streptomyces lividans, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-01-05 Classification: IMMUNE SYSTEM/TRANSPORT PROTEIN Ligands: K |
 |
Organism: Streptomyces lividans, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-01-05 Classification: IMMUNE SYSTEM/TRANSPORT PROTEIN Ligands: K |
 |
Organism: Streptomyces lividans, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2010-07-14 Classification: membrane protein/metal transport Ligands: RB |
 |
Organism: Mus musculus, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2010-06-23 Classification: IMMUNE SYSTEM/metal TRANSPORT Ligands: K |
 |
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-10-09 Classification: VIRAL PROTEIN Ligands: ZN, NA |
 |
Structure Of Hepatitis C Viral Ns3 Protease Domain Complexed With Ns4A Peptide And Ketoamide Sch476776
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-07-31 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, BME, HUD |

