Search Count: 20
 |
Crystal Structure Of Cim-2, A Membrane-Bound B1 Metallo-Beta-Lactamase From Chryseobacterium Indologenes
Organism: Chryseobacterium indologenes
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: ANTIMICROBIAL PROTEIN Ligands: ZN, MPD, CL, IPA, PGR |
 |
Crystal Structure Of Cjo-1, A Membrane-Bound B1 Metallo-Beta-Lactamase From Chryseobacterium Joostei
Organism: Chryseobacterium joostei
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: ANTIMICROBIAL PROTEIN Ligands: GOL, ZN |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: AV0, AGS, MG |
 |
Cryo-Em Structure Of S. Aureus Targh In Complex With Amp-Pnp And Targocil-Ii
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: A1AV9, ANP, MG |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: AV0, AGS, MG |
 |
Cryo-Em Structure Of S. Aureus Targh In Complex With Targocil-Ii And Atp-Gamma-S In A Catalytically Competent Conformation
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: A1AV9, MG, AGS |
 |
Cryo-Em Structure Of S. Aureus Targh In Complex With Targocil-Ii And Atp-Gamma-S In A Catalytically Incompetent Conformation
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: AV0, A1AV9, AGS, MG |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: RIBOSOME Ligands: GNP |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: RIBOSOME Ligands: GNP |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: RIBOSOME Ligands: GNP |
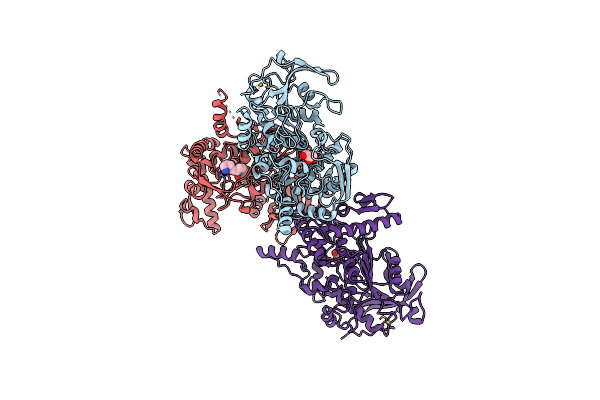 |
Organism: Fusobacterium nucleatum subsp. nucleatum atcc 25586
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-03-24 Classification: HYDROLASE Ligands: GOL, MG, SPD |
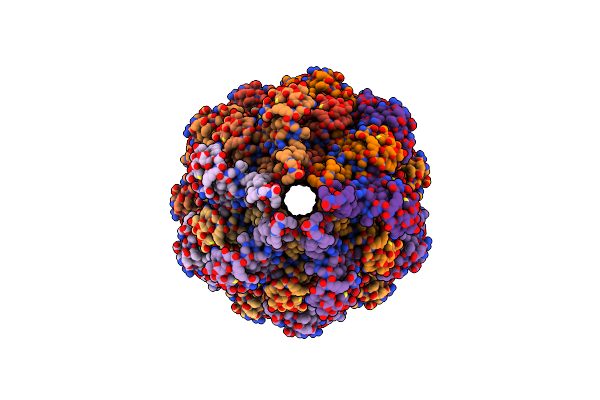 |
Organism: Escherichia coli o127:h6 (strain e2348/69 / epec)
Method: ELECTRON MICROSCOPY Release Date: 2020-12-30 Classification: PROTEIN TRANSPORT |
 |
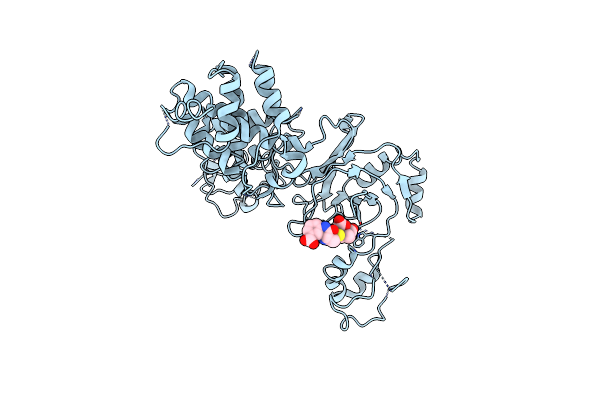
Organism: Citrobacter rodentium (strain icc168)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-11-25 Classification: TRANSFERASE Ligands: 1RG |
 |
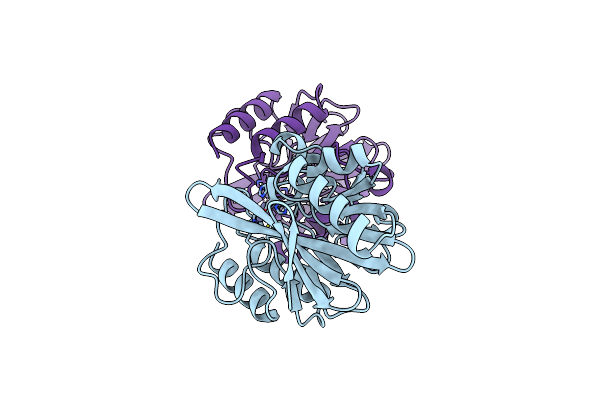
Organism: Salmonella enterica
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2020-11-25 Classification: TRANSFERASE Ligands: 1RG |
 |
Crystal Structure Of A Chimeric Ndm-1 Metallo-Beta-Lactamase Harboring The Imp-1 L3 Loop
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of Ndm-1 Metallo-Beta-Lactamase Harboring An Insertion Of A Pro Residue In L3 Loop
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: ZN, CD, NI, CO, CA, SO4 |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2012-12-26 Classification: GENE REGULATION Ligands: ZN |
 |
The Crystal Structure Of The Ing4 Dimerization Domain Reveals The Functional Organization Of The Ing Family Of Chromatin Binding Proteins.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2012-02-22 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2008-04-15 Classification: GENE REGULATION Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2008-04-01 Classification: CELL CYCLE |

