Search Count: 85
 |
Organism: Red-crowned crane parvovirus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: VIRUS Ligands: MG |
 |
Organism: Turkey parvovirus 260
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: VIRUS Ligands: MG |
 |
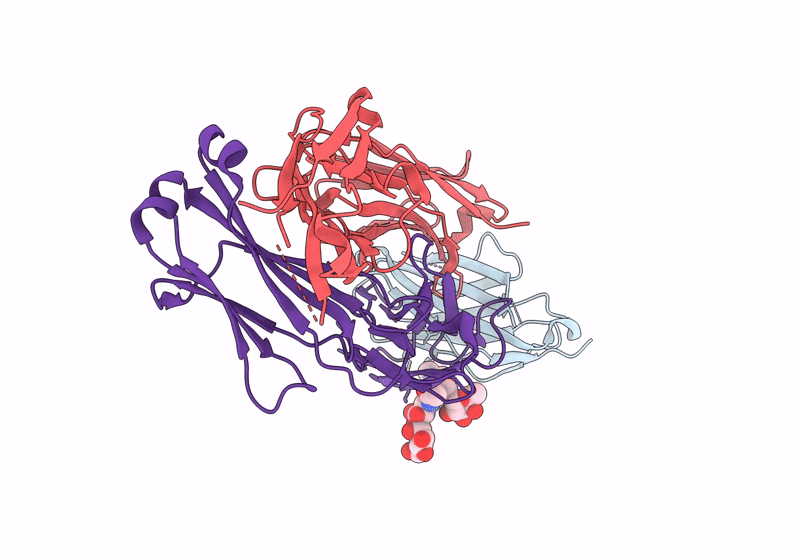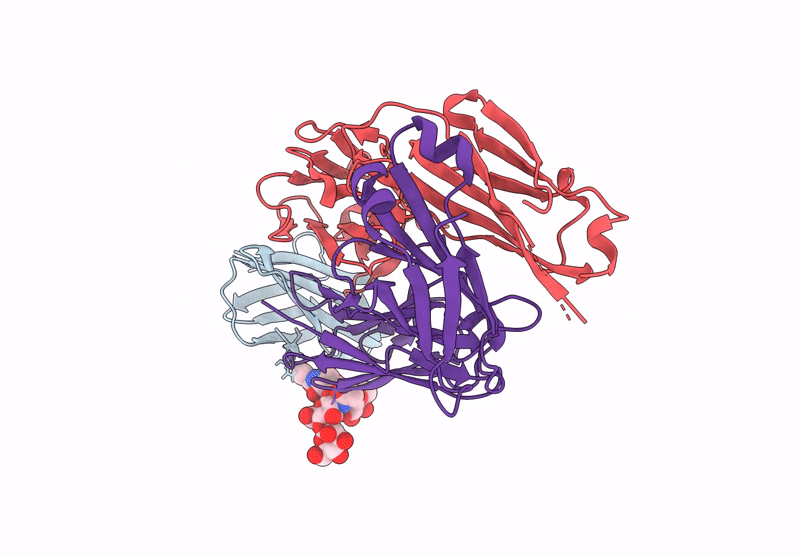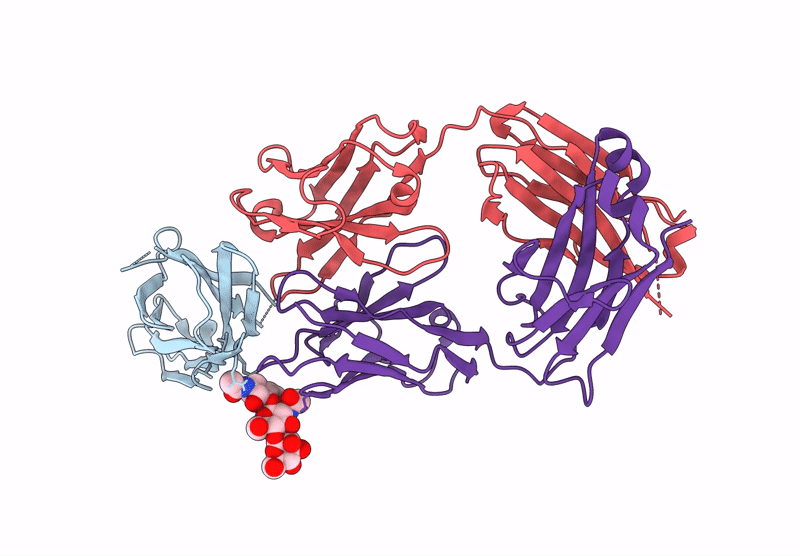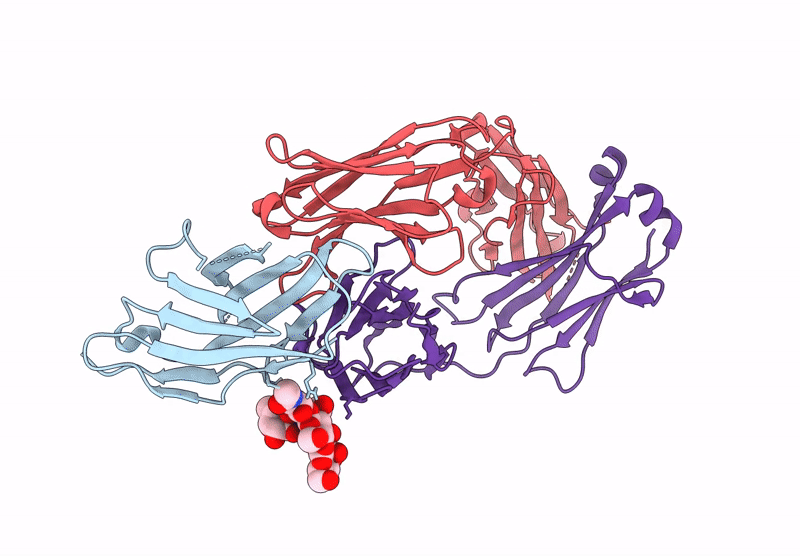
Crystal Structure Of Engineered Ipilimumab (Mipi.4) Fab In Complex With Human Ctla-4
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-02-05 Classification: ANTITUMOR PROTEIN/IMMUNE SYSTEM |
 |
Crystal Structure Of Engineered Ipilimumab (Mipi.4) Fab In Complex With Mouse Ctla-4
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2025-02-05 Classification: ANTITUMOR PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-02-05 Classification: ANTITUMOR PROTEIN/IMMUNE SYSTEM |
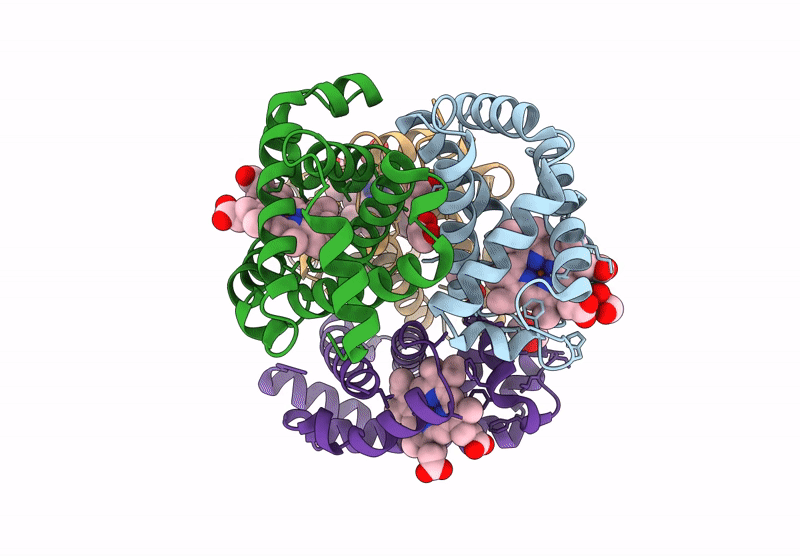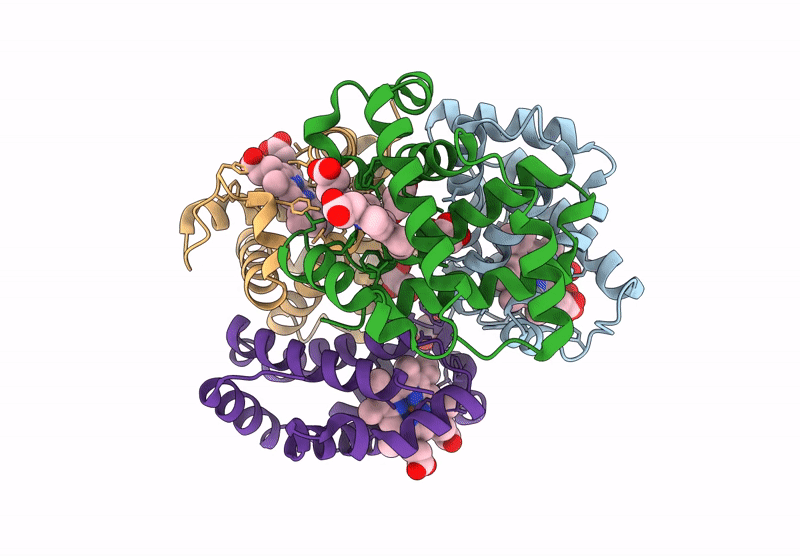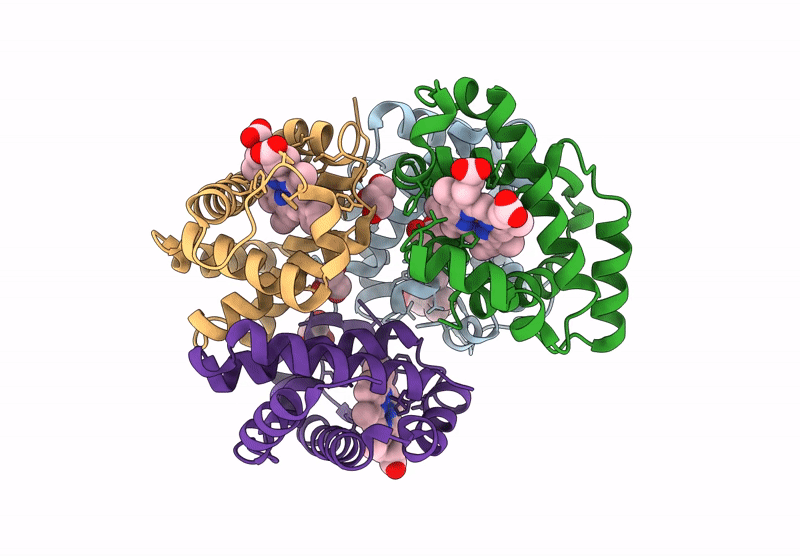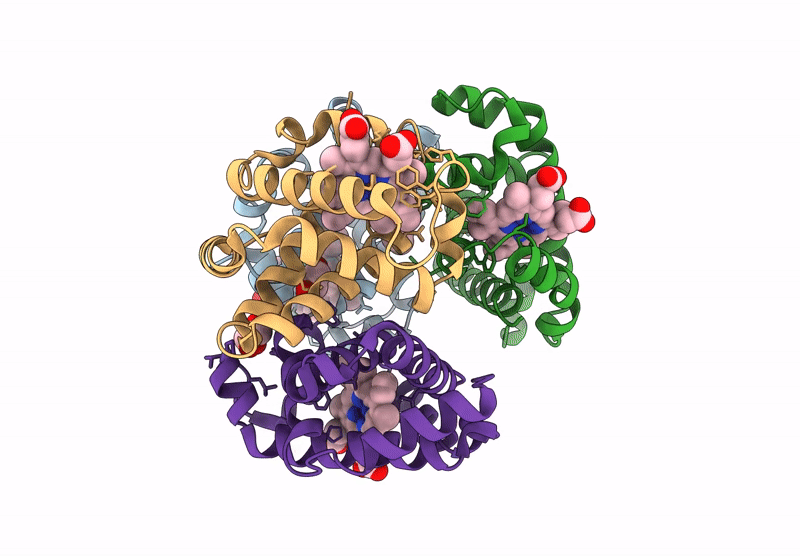 |
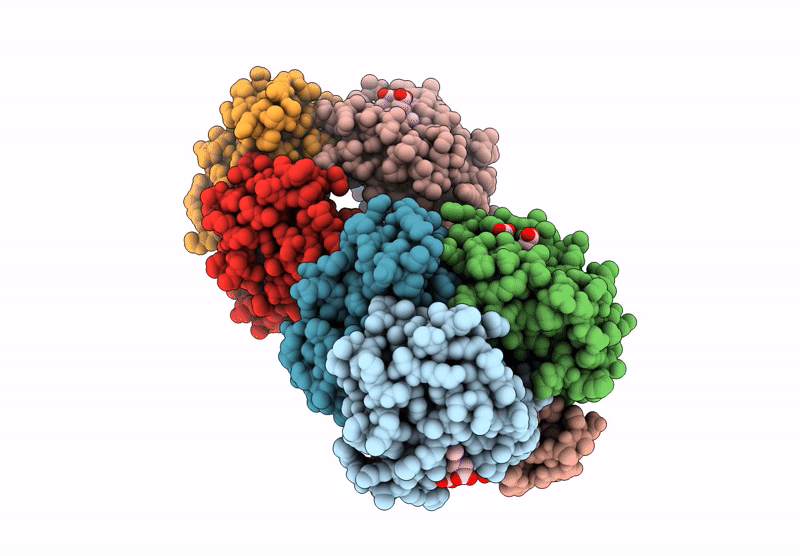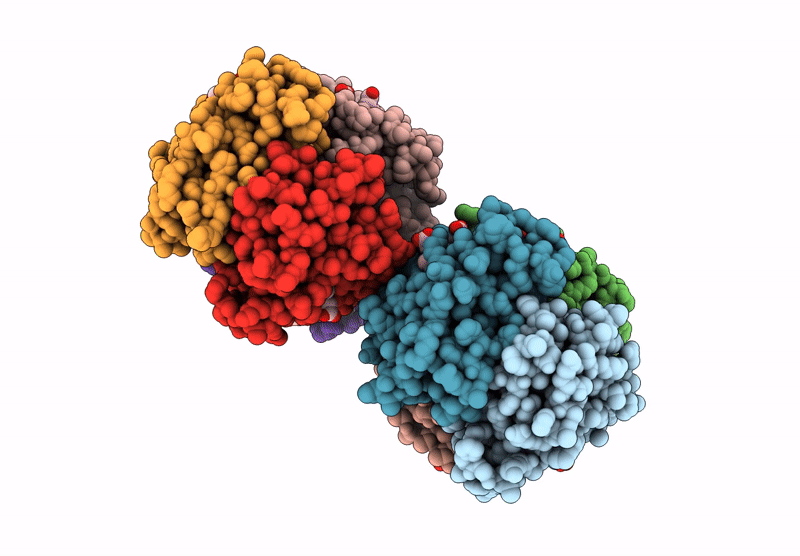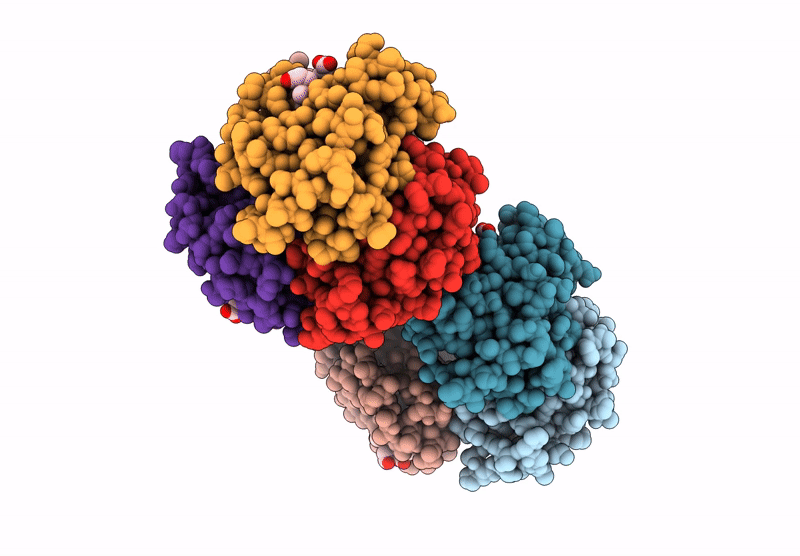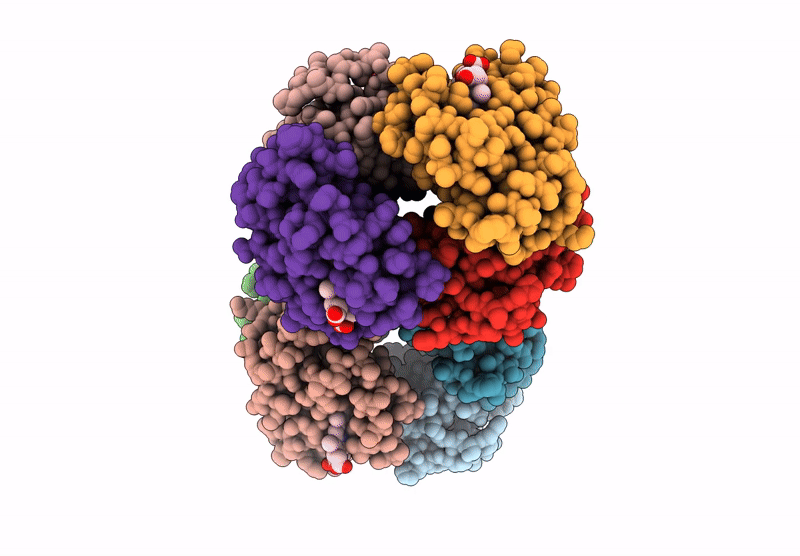
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-12-11 Classification: OXYGEN TRANSPORT Ligands: GOL, HEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2024-12-11 Classification: OXYGEN TRANSPORT Ligands: HEM |
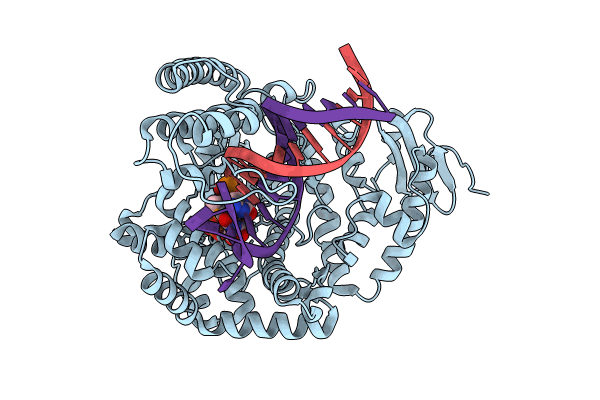 |
Klentaq Dna Polymerase In A Ternary Complex With Primer/Template And A Selenophene-Modified Dutp (Sedutp)
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2024-11-13 Classification: TRANSFERASE Ligands: A1IDZ, MG, DOC |
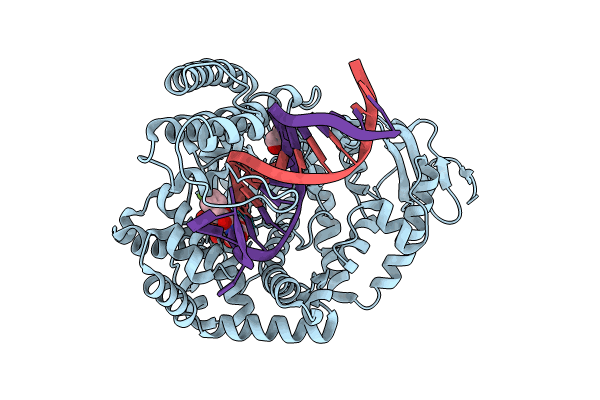 |
Klentaq Dna Polymerase In A Ternary Complex With Primer/Template And A Fluorobenzofuran-Modified Dutp (Fbfdutp)
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-11-13 Classification: TRANSFERASE Ligands: MG, A1IDW, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2024-07-17 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
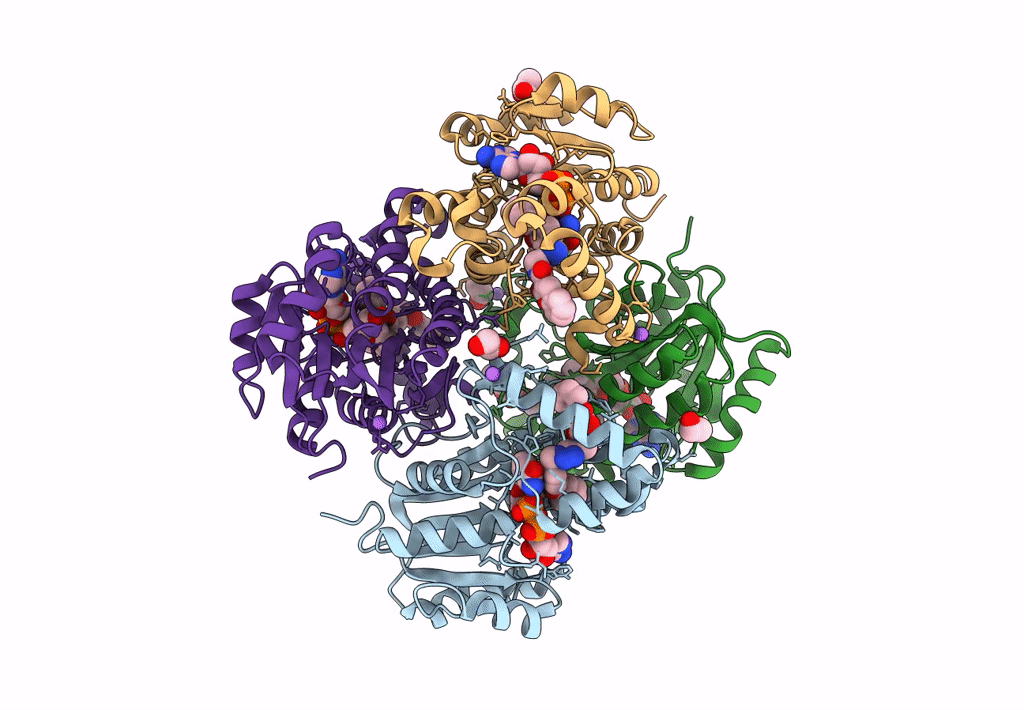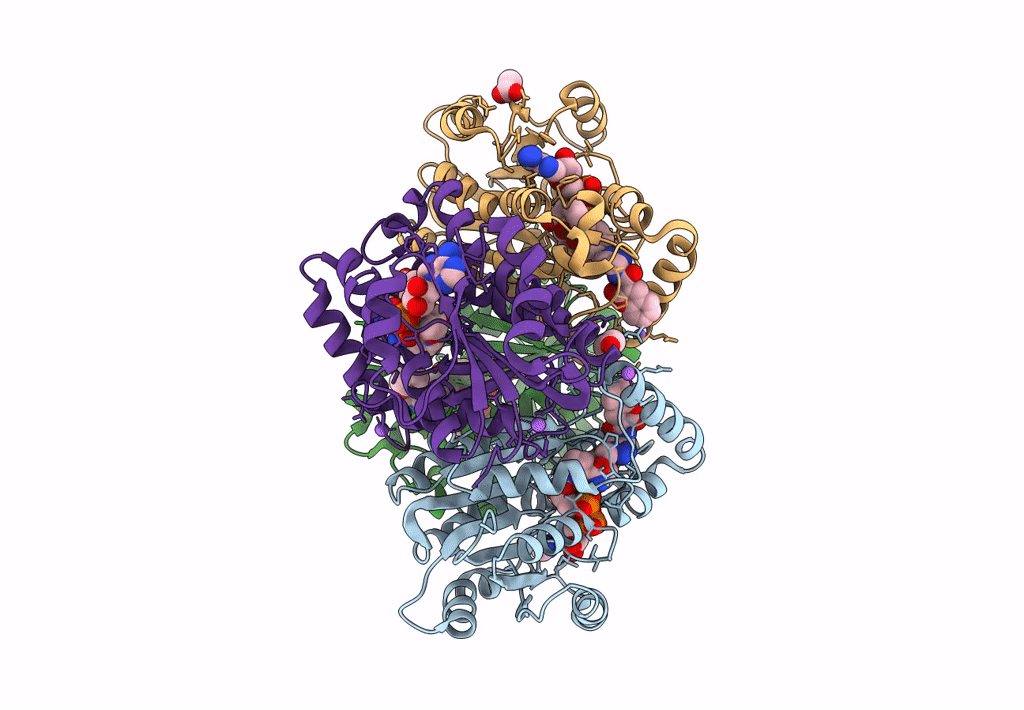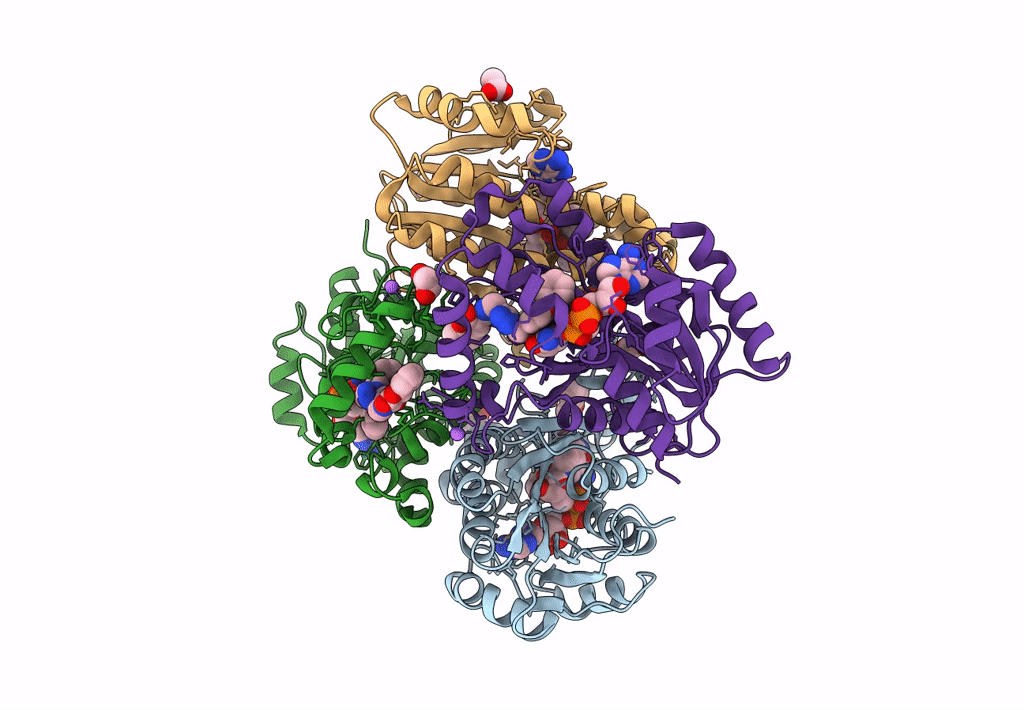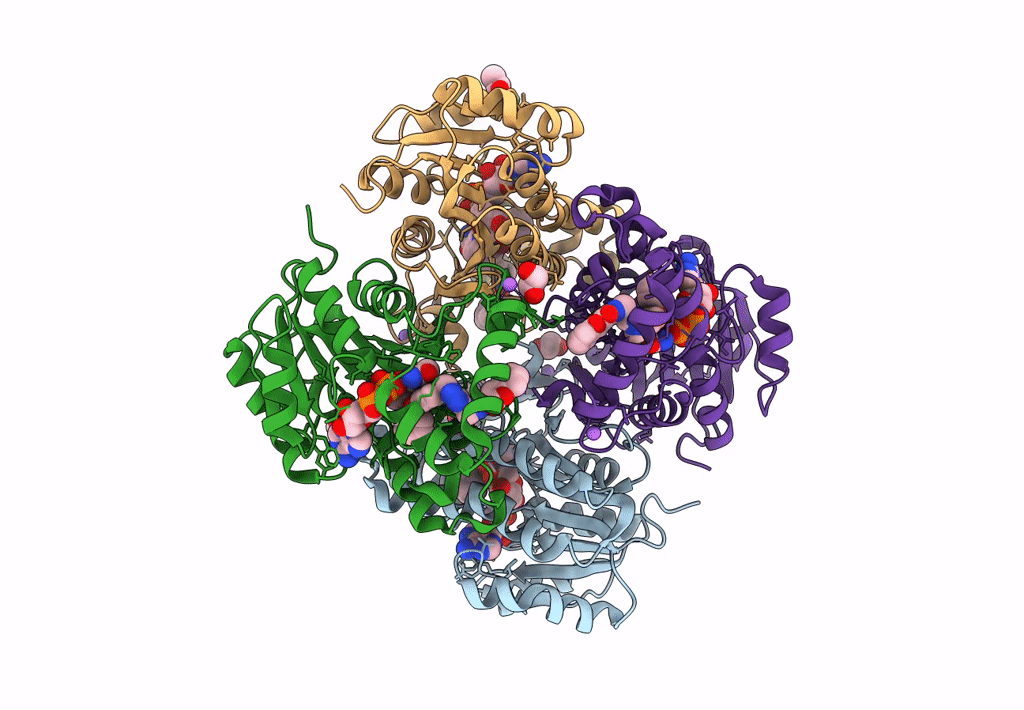
Crystal Structure Of Heterotrimeric Anti-Tigit Fabs In Complex With Human Tigit
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2023-09-20 Classification: IMMUNE SYSTEM,PROTEIN BINDING Ligands: NAG |
 |
Structure Of Inha From Mycobacterium Tuberculosis In Complex With N-((1-(3-Hydroxy-4-Phenoxybenzyl)-1H-1,2,3-Triazol-4-Yl)Methyl)-2-Oxo-2H-Chromene-3-Carboxamide
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-08-16 Classification: OXIDOREDUCTASE Ligands: NAD, VZI, EDO, NA, ACT |
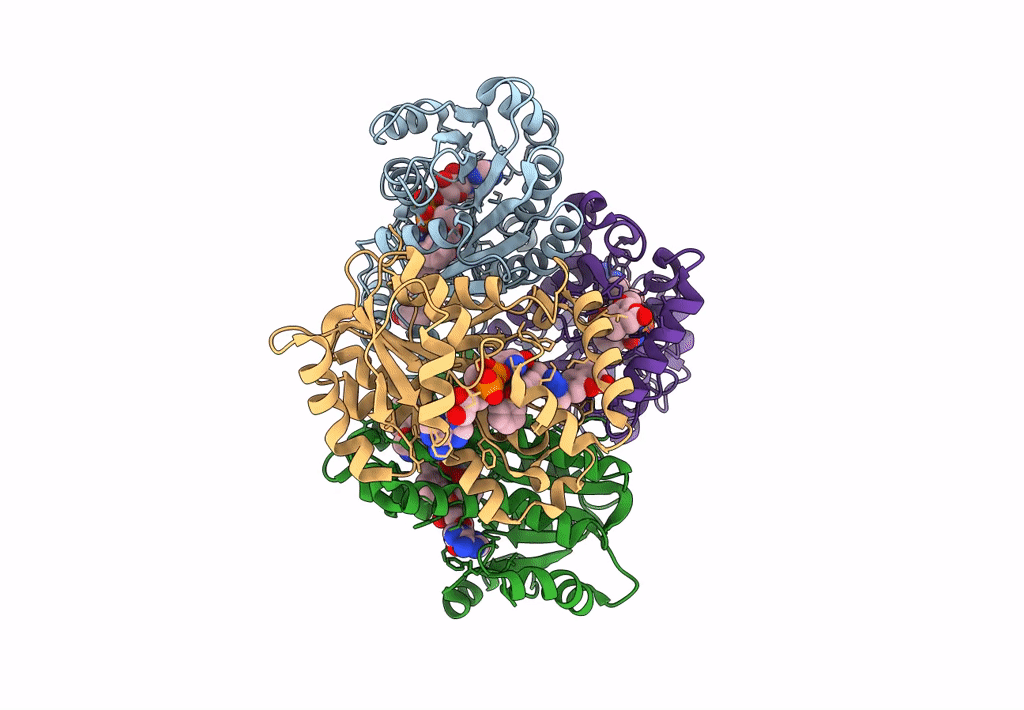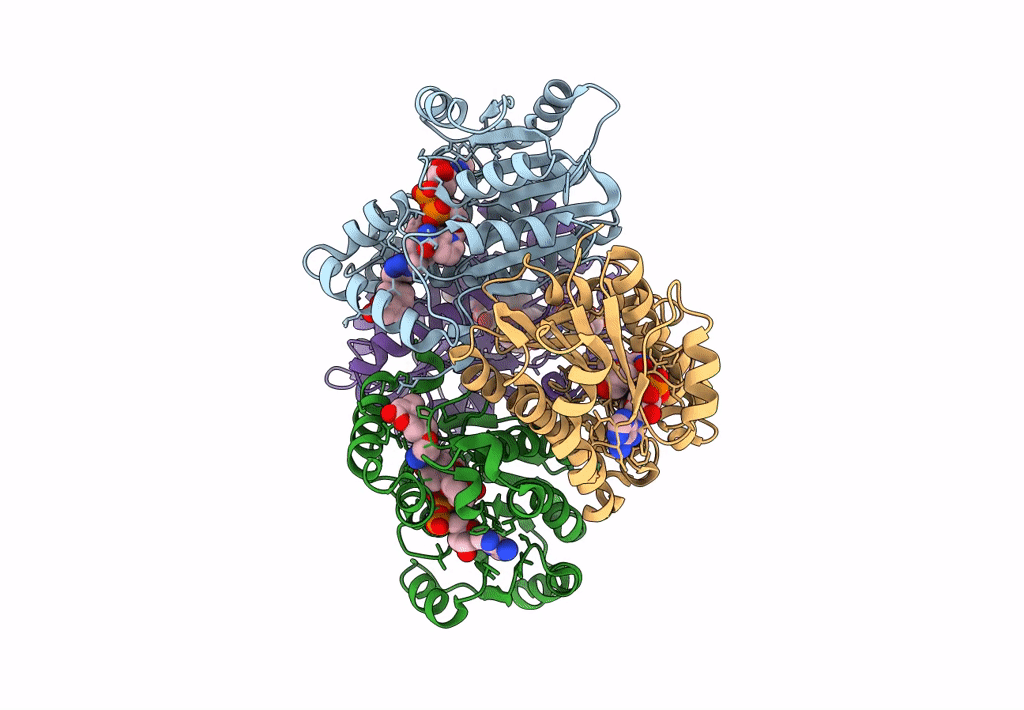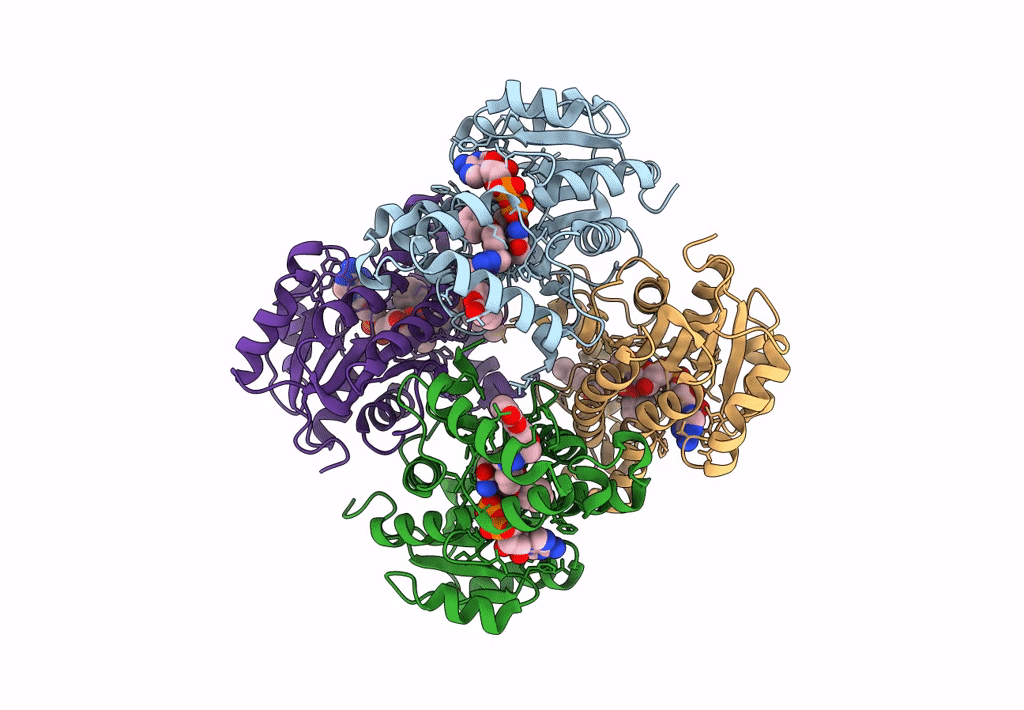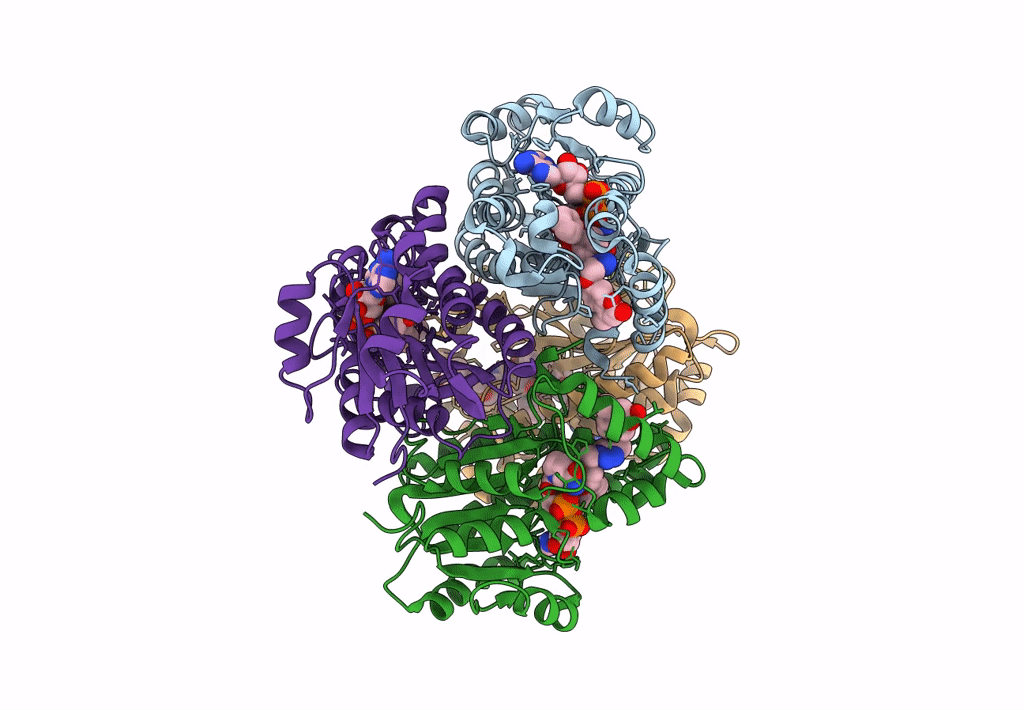 |
Structure Of Inha From Mycobacterium Tuberculosis In Complex With Inhibitor 7-((1-(3-Hydroxy-4-Phenoxybenzyl)-1H-1,2,3-Triazol-4-Yl)Methoxy)-4-Methyl-2H-Chromen-2-One
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2023-08-16 Classification: OXIDOREDUCTASE Ligands: NAD, VZR |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Barbituric Acid And Citric Acid, Form I
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2023-07-12 Classification: HYDROLASE Ligands: BR8, CIT, EDO, NA |
 |
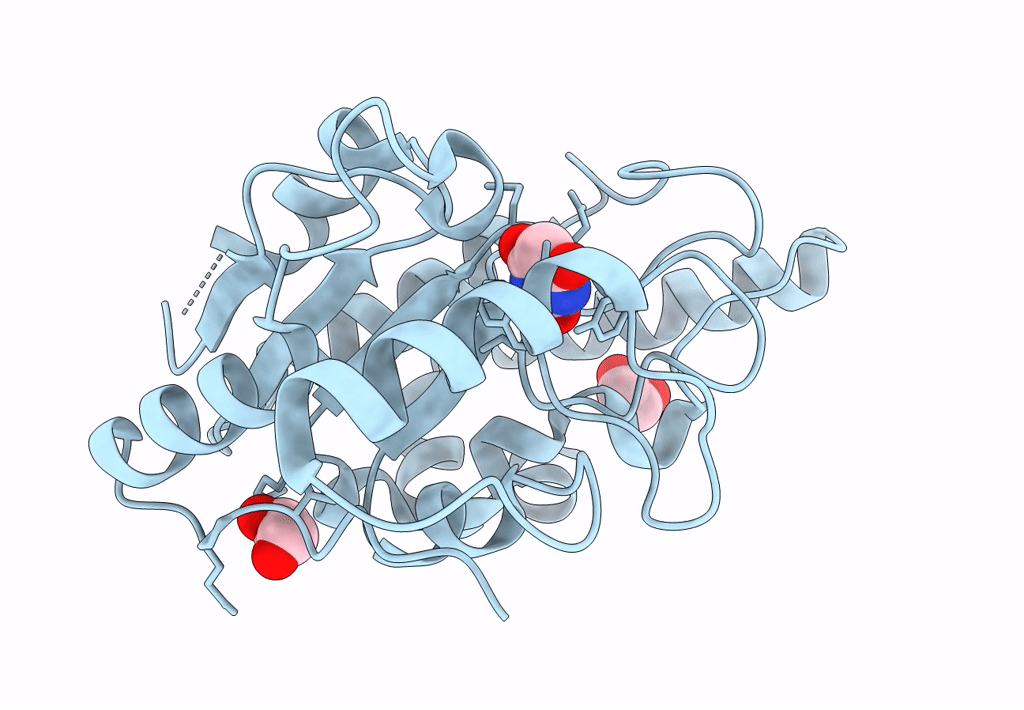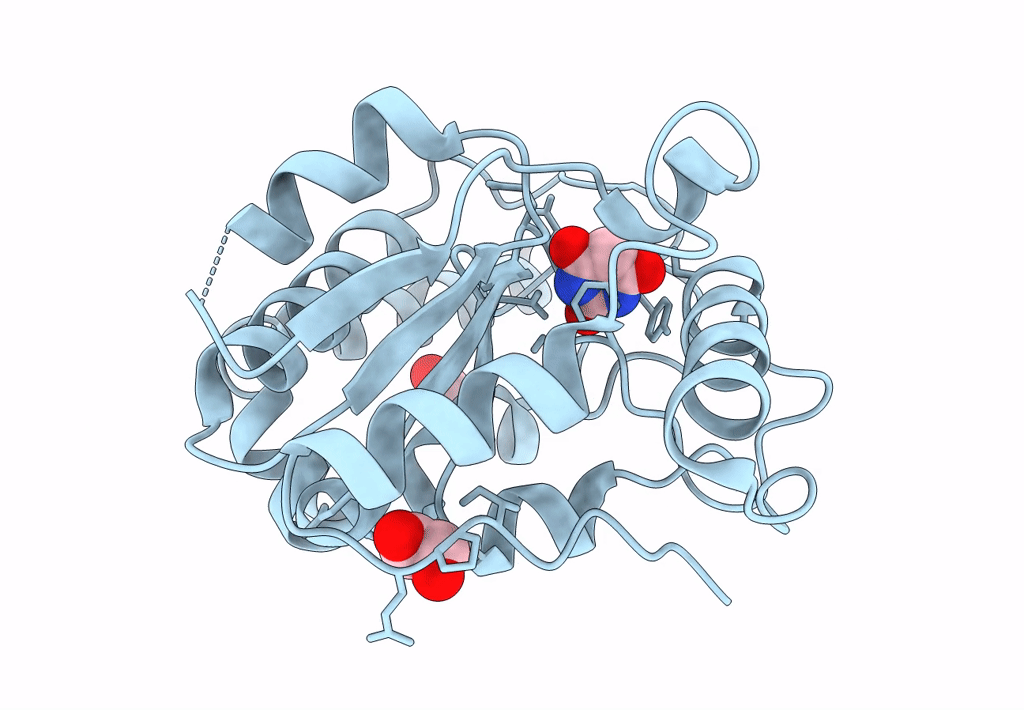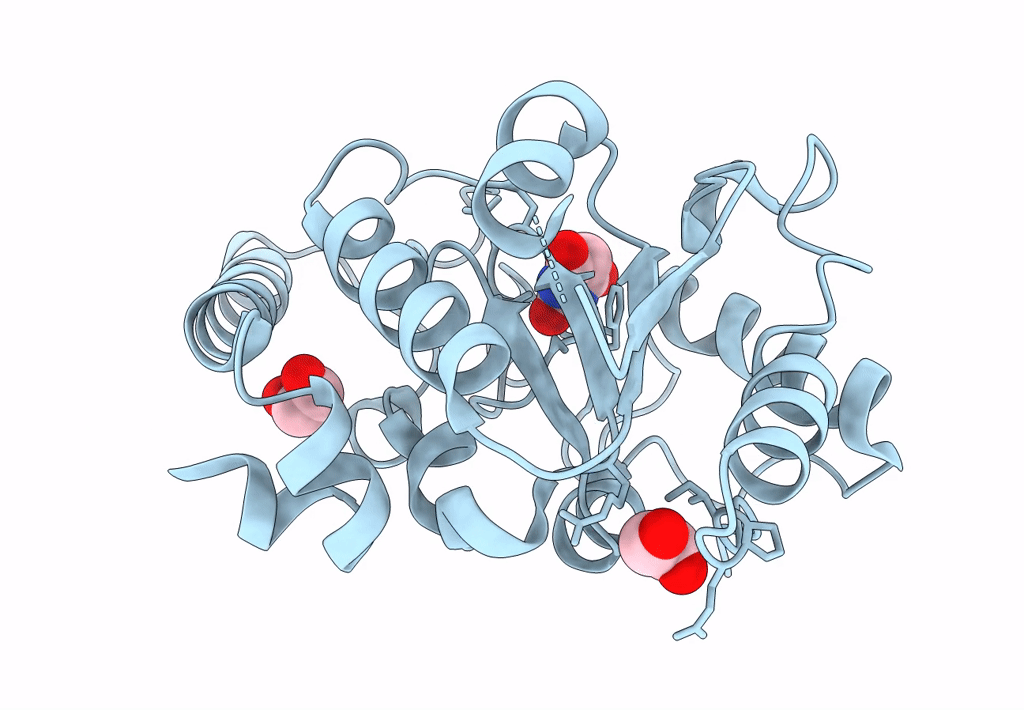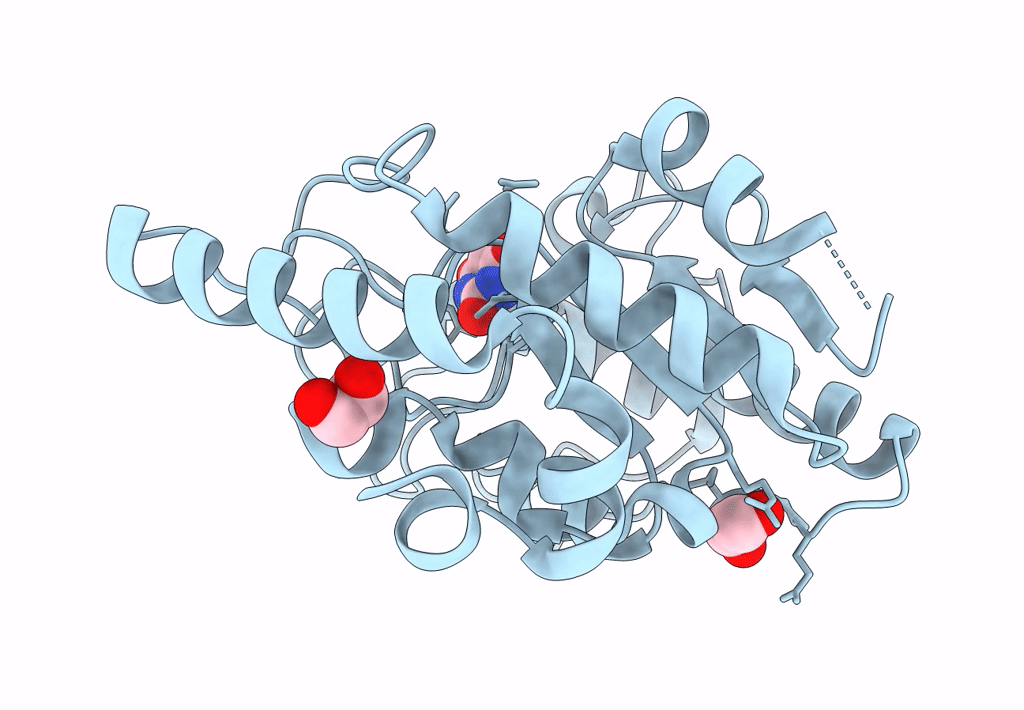
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Barbituric Acid, Form I
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: BR8, EDO, CL |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Barbituric Acid, Form Iii
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: BR8, EDO |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Barbituric Acid, Form Ii
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: BR8, EDO |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Isoorotic Acid (2,4-Dihydroxypyrimidine-5-Carboxylic Acid), Form I
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: 5CU, EDO |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Isoorotic Acid (2,4-Dihydroxypyrimidine-5-Carboxylic Acid) And Citric Acid, Form I
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: 5CU, CIT |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With 2,4-Thiazolidinedione, Form I
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: OD3, EDO |

