Search Count: 20
 |
Crystal Structure Of The Cbm3 From Bacillus Subtilis At 1.06 Angstrom Resolution
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: PEG |
 |
Crystal Structure Of The Cbm3 From Bacillus Subtilis At 1.28 Angstrom Resolution
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2020-09-30 Classification: HYDROLASE |
 |
Crystal Structure Of The Endo-Beta-1,4-Glucanase (Xac0030) From Xanthomonas Axonopodis Pv. Citri With The Triple Mutation His174Trp, Tyr211Ala And Lys227Arg.
Organism: Xanthomonas axonopodis pv. citri
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2017-01-25 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of The Endo-Beta-1,4-Glucanase Xac0029 From Xanthomonas Axonopodis Pv. Citri
Organism: Xanthomonas axonopodis pv. citri
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-01-25 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Xanthomonas axonopodis pv. citri
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-01-25 Classification: HYDROLASE |
 |
Organism: Xanthomonas axonopodis pv. citri
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2015-10-21 Classification: HYDROLASE Ligands: CAC |
 |
Organism: Xanthomonas axonopodis pv. citri
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2015-10-21 Classification: HYDROLASE |
 |
Organism: Xanthomonas axonopodis pv. citri
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2015-10-21 Classification: HYDROLASE |
 |
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-02-05 Classification: HYDROLASE Ligands: TRS |
 |
Native Structure Of Endo-1,4-Beta-D-Mannanase From Thermotoga Petrophila Rku-1
Organism: Thermotoga petrophila
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2011-12-28 Classification: HYDROLASE |
 |
I222 Crystal Form Of The Hyperthermostable Endo-1,4-Beta-D-Mannanase From Thermotoga Petrophila Rku-1
Organism: Thermotoga petrophila
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2011-12-28 Classification: HYDROLASE Ligands: TRS, GOL, PO4 |
 |
Structure Of The Hyperthermostable Endo-1,4-Beta-D-Mannanase From Thermotoga Petrophila Rku-1 In Complex With Beta-D-Glucose
Organism: Thermotoga petrophila
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2011-12-28 Classification: HYDROLASE Ligands: BGC |
 |
Structure Of The Hyperthermostable Endo-1,4-Beta-D-Mannanase From Thermotoga Petrophila Rku-1 With Three Glycerol Molecules
Organism: Thermotoga petrophila rku-1
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2011-12-28 Classification: HYDROLASE Ligands: GOL, TRS |
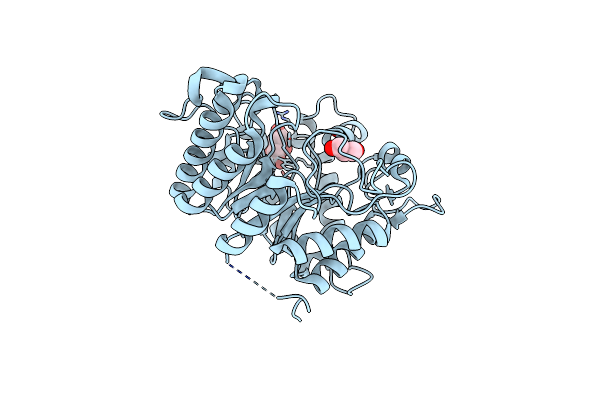 |
Structure Of The Hyperthermostable Endo-1,4-Beta-D-Mannanase From Thermotoga Petrophila Rku-1 With Citrate And Glycerol
Organism: Thermotoga petrophila rku-1
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2011-12-28 Classification: HYDROLASE Ligands: CIT, GOL |
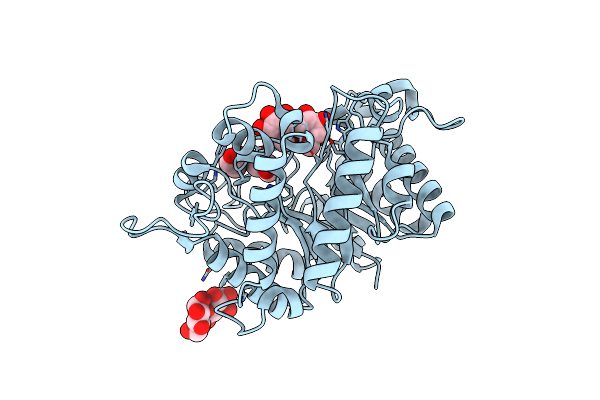 |
Structure Of The Hyperthermostable Endo-1,4-Beta-D-Mannanase From Thermotoga Petrophila Rku-1 In Complex With Three Maltose Molecules
Organism: Thermotoga petrophila rku-1
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2011-12-28 Classification: HYDROLASE Ligands: GOL |
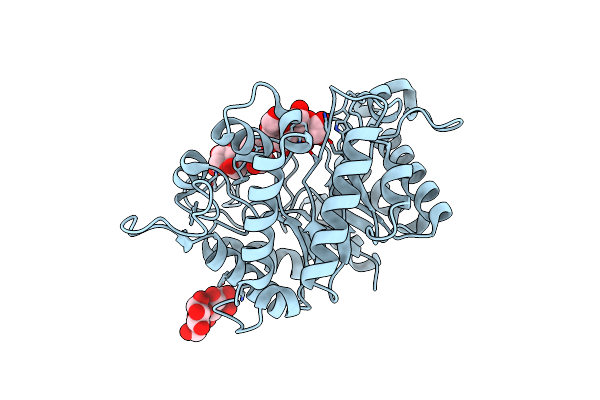 |
Structure Of The Hyperthermostable Endo-1,4-Beta-D-Mannanase From Thermotoga Petrophila Rku-1 With Maltose And Glycerol
Organism: Thermotoga petrophila rku-1
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2011-12-28 Classification: HYDROLASE |
 |
 |
Structure Of The Endo-1,4-Beta-Glucanase From Bacillus Subtilis 168 With Manganese(Ii) Ion
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2011-09-14 Classification: HYDROLASE Ligands: MN, GOL, PO4 |
 |
P212121 Crystal Form Of The Endo-1,4-Beta-Glucanase From Bacillus Subtilis 168
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-09-14 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2011-09-14 Classification: HYDROLASE |

