Search Count: 57
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-02-07 Classification: OXIDOREDUCTASE Ligands: BME, FMN, PLP, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2024-02-07 Classification: OXIDOREDUCTASE Ligands: PO4, FMN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: TRANSFERASE Ligands: PLP |
 |
Structure Of A Delta-N Mutant - E232Start - Of Pa1120 (Tpbb Or Yfin) From Pseudomonas Aeruginosa (Pao1) Comprising Only The Ggdef Domain
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-07-07 Classification: LIGASE Ligands: MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-06-03 Classification: TRANSFERASE Ligands: NI, EDO, O3H |
 |
Crystal Structure Of The Complex Between Ppargamma Lbd And The Ligand Nv1346 (3A)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-02-26 Classification: TRANSCRIPTION Ligands: MX8 |
 |
Crystal Structure Of Human R180T Variant Of Ornithine Aminotransferase At 1.8 Angstrom
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-06-05 Classification: TRANSFERASE Ligands: PLP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2018-10-10 Classification: TRANSFERASE Ligands: PLP, CL |
 |
Structure Of The Hybrid Domain (Ggdef-Eal) Of Pa0575 From Pseudomonas Aeruginosa Pao1 At 2.8 Ang. With Gtp And Ca2+ Bound To The Active Site Of The Ggdef Domain
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-12-20 Classification: HYDROLASE Ligands: CA, GTP |
 |
Crystal Structure Of The D183N Variant Of Human Alanine:Glyoxylate Aminotransferase Major Allele (Agt-Ma) At Ph 9.0. 2.8 Ang; Internal Aldimine With Plp In The Active Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-09-27 Classification: TRANSFERASE Ligands: PLP, DIO |
 |
Crystal Structure Of Human Alanine:Glyoxylate Aminotransferase Major Allele (Agt-Ma) At 2.5 Angstrom; Internal Aldimine With Plp In The Active Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-09-27 Classification: TRANSFERASE Ligands: PLP |
 |
Crystal Structure Of The D183N Variant Of Human Alanine:Glyoxylate Aminotransferase Major Allele (Agt-Ma) At 1.8 Angstrom; Internal Aldimine With Plp In The Active Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-09-20 Classification: TRANSFERASE Ligands: PLP, BTB |
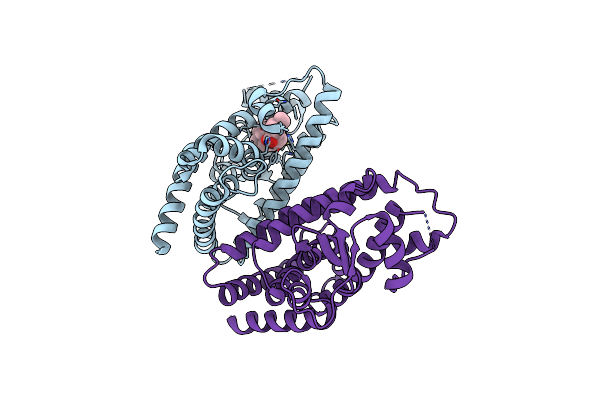 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-08-09 Classification: SIGNALING PROTEIN Ligands: QZQ |
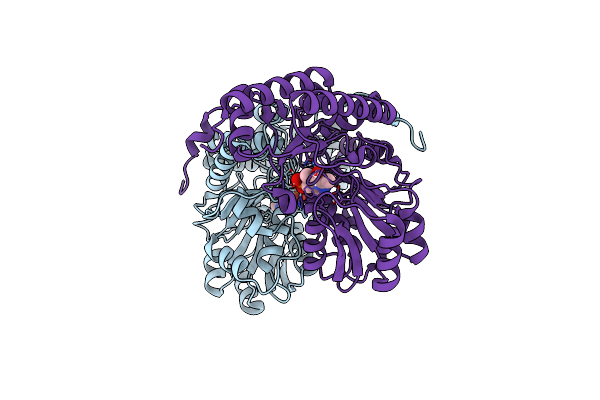 |
Structure Of Human Alanine:Glyoxylate Aminotransferase Major Allele (Agt-Ma) Showing X-Ray Induced Reduction Of Plp Internal Aldimine To 4'-Deoxy-Piridoxine-Phosphate (Plr)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-01-25 Classification: TRANSFERASE Ligands: PLR |
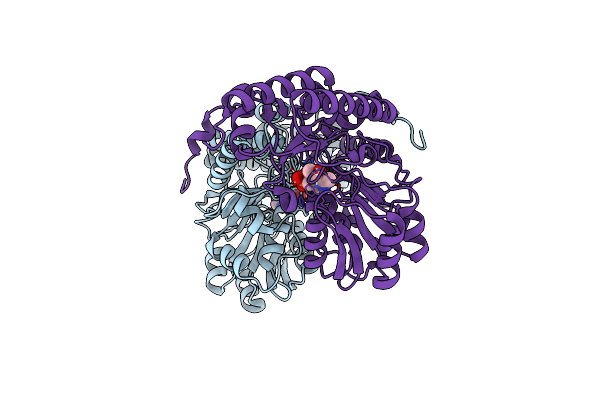 |
Crystal Structure Of Human Alanine:Glyoxylate Aminotransferase Major Allele (Agt-Ma) At 1.7 Angstrom; Internal Aldimine With Plp In The Active Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-12-21 Classification: TRANSFERASE Ligands: PLP |
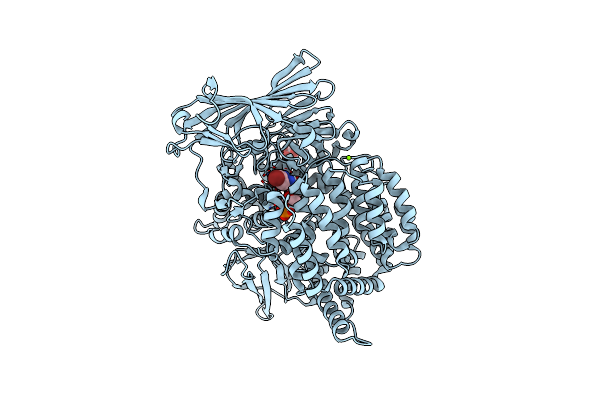 |
X-Ray Crystal Structure Of Pfa-M1 In Complex With Hydroxamic Acid-Based Inhibitor 9B
Organism: Plasmodium falciparum (isolate fcb1 / columbia)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-03-30 Classification: HYDROLASE/HYDROLASE inhibitor Ligands: 4S9, ZN, GOL, MG, PO4 |
 |
X-Ray Crystal Structure Of Pfa-M1 In Complex With Hydroxamic Acid-Based Inhibitor 9F
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-03-30 Classification: HYDROLASE/HYDROLASE inhibitor Ligands: ZN, 4SA, MG |
 |
X-Ray Crystal Structure Of Pfa-M1 In Complex With Hydroxamic Acid-Based Inhibitor 9Q
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-03-30 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 4SY, ZN, MG, GOL, DMS, PO4 |
 |
X-Ray Crystal Structure Of Pfa-M1 In Complex With Hydroxamic Acid-Based Inhibitor 9M
Organism: Plasmodium falciparum (isolate fcb1 / columbia)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-03-30 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, 4SZ, MG, GOL, DMS |

