Search Count: 15
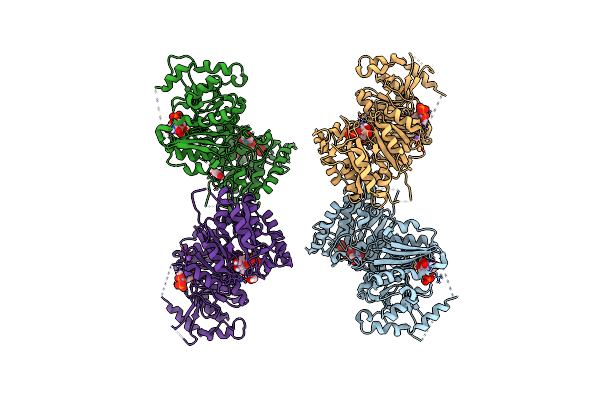 |
Crystal Structure Of Heparosan Synthase 2 From Pasteurella Multocida At 2.85 A
Organism: Pasteurella multocida
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2024-09-04 Classification: TRANSFERASE Ligands: MN, UDP, NA, CL, EDO, CA |
 |
Crystal Structure Of Heparosan Synthase 2 From Pasteurella Multocida At 1.98 A
Organism: Pasteurella multocida
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2024-07-24 Classification: TRANSFERASE Ligands: MN, UDP, EDO, NA |
 |
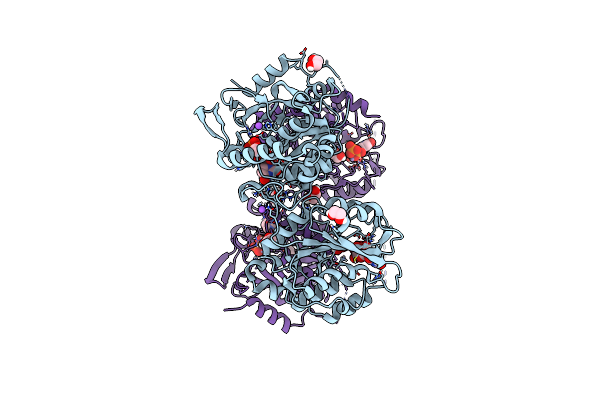
Cryo-Em Structure Of Heparosan Synthase 2 From Pasteurella Multocida With Polysaccharide In The Glcnac-T Active Site
Organism: Pasteurella multocida
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: TRANSFERASE Ligands: MN, UDP |
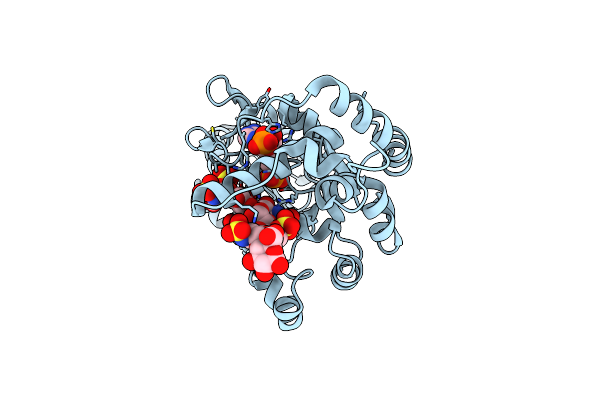 |
Ternary Complex Of Fixed-Arm Trx-3Ost5 (I299E) With 8Mer-1 Octasaccharide Substrate And Co-Factor Product Pap
Organism: Escherichia coli, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-01-19 Classification: TRANSFERASE/SUBSTRATE Ligands: A3P |
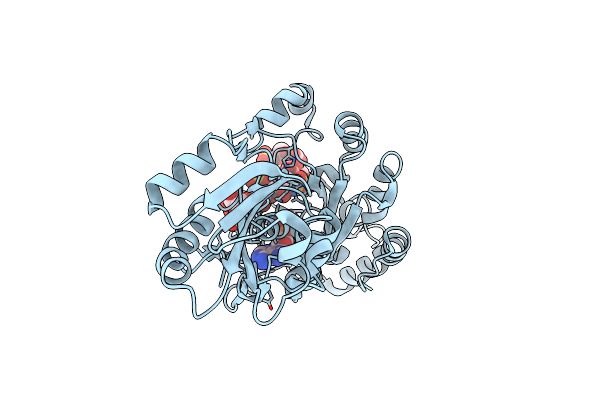 |
Ternary Complex Of Fixed-Arm Trx-3Ost5 (I299E) With 8Mer-2 Octasaccharide Substrate And Co-Factor Product Pap
Organism: Escherichia coli, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2022-01-19 Classification: TRANSFERASE/SUBSTRATE Ligands: A3P |
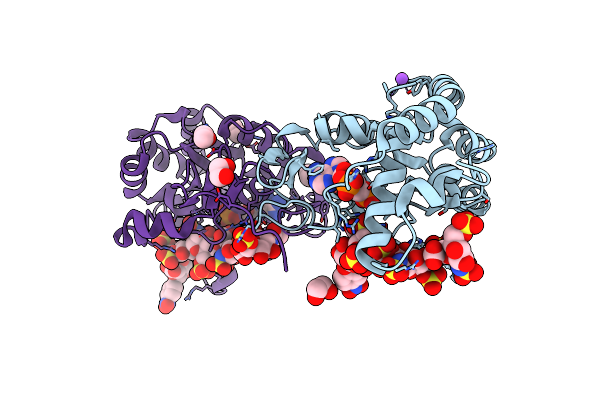 |
Crystal Structure Of 3-O-Sulfotransferase Isoform 3 In Complex With 8Mer Oligosaccharide With 6S Sulfation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2021-06-23 Classification: TRANSFERASE Ligands: NPO, A3P, NA, EDO |
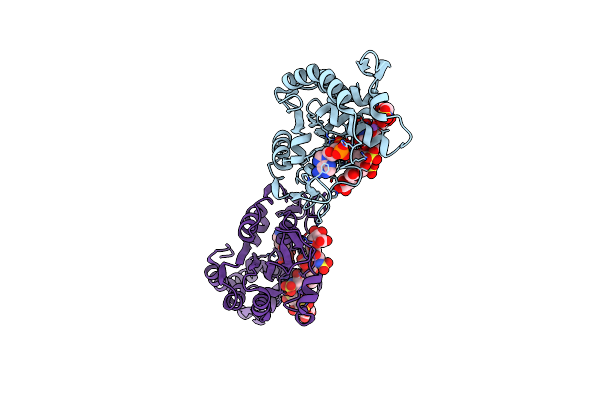 |
Crystal Structure Of 3-O-Sulfotransferase Isoform 3 In Complex With 8Mer Oligosaccharide With No 6S Sulfation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2021-06-23 Classification: TRANSFERASE |
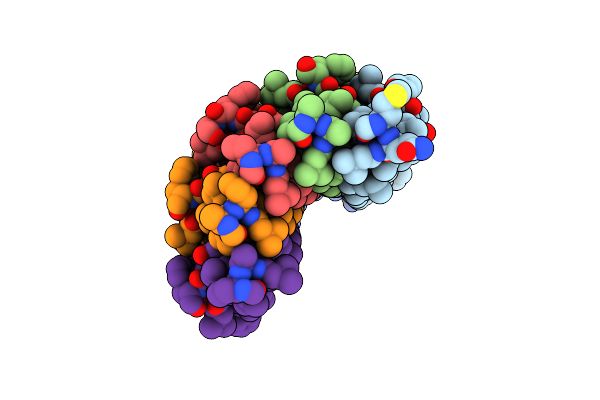 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-02-08 Classification: MEMBRANE PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-02-08 Classification: MEMBRANE PROTEIN |
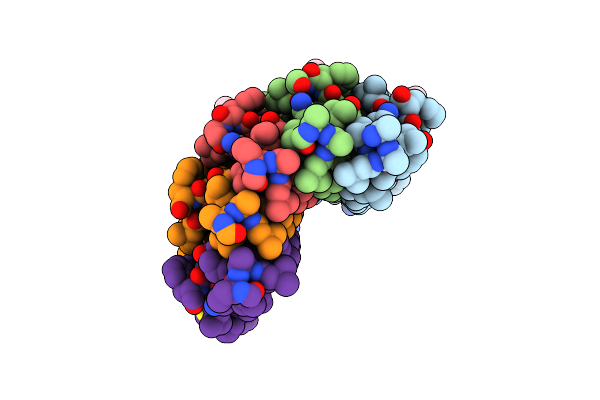 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-02-08 Classification: MEMBRANE PROTEIN Ligands: DCW |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-02-08 Classification: MEMBRANE PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2010-09-15 Classification: HYDROLASE Ligands: ANP, MG, PO4 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2010-09-15 Classification: HYDROLASE Ligands: ANP, MG, PO4 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2010-09-15 Classification: HYDROLASE Ligands: ANP, MG |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2010-09-15 Classification: HYDROLASE Ligands: ANP, MG |

