Search Count: 50
 |
Structure Of Paenibacillus Polymyxa Gs Bound To Met-Sox-P-Adp (Transition State Complex) To 1.98 Angstom
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2022-06-29 Classification: LIGASE Ligands: ADP, P3S, MG |
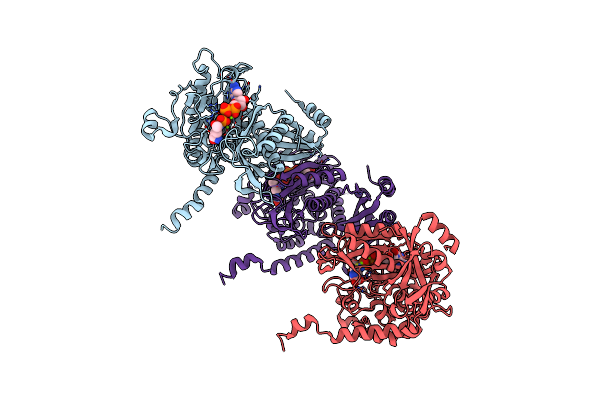
 |
Organism: Paenibacillus polymyxa
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN, LIGASE Ligands: MG, GLN |
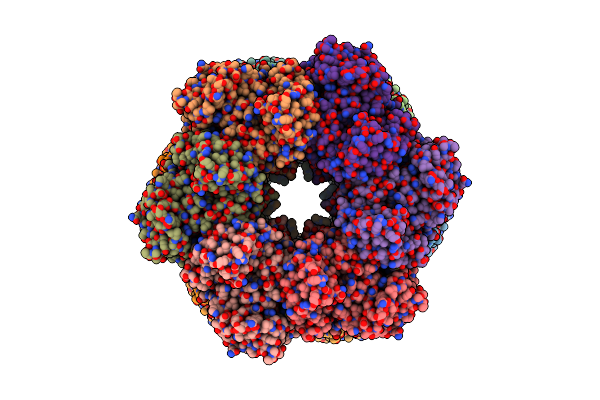
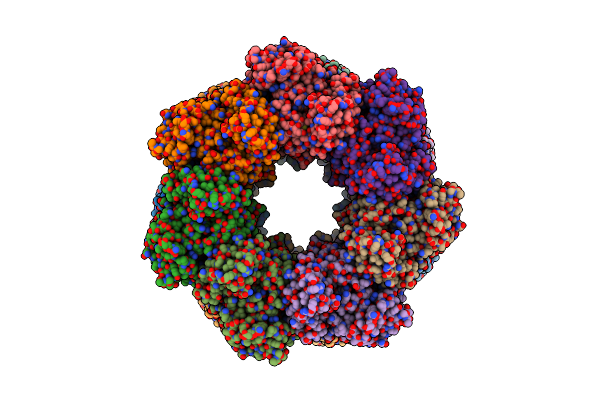 |
Organism: Paenibacillus polymyxa
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN, LIGASE Ligands: MG, GLN |
 |
Organism: Paenibacillus polymyxa
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN Ligands: MG |
 |
Structure Of Beta-Glucosidase A From Paenibacillus Polymyxa Complexed With Multivalent Inhibitors.
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2019-07-03 Classification: HYDROLASE Ligands: JJW |
 |
Structure Of Beta-Glucosidase A From Paenibacillus Polymyxa Complexed With A Monovalent Inhibitor
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2019-07-03 Classification: HYDROLASE Ligands: JSK |
 |
Organism: Homo sapiens, Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2016-09-21 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Homo sapiens, Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2016-09-21 Classification: OXIDOREDUCTASE/REPRESSOR Ligands: FAD, NA |
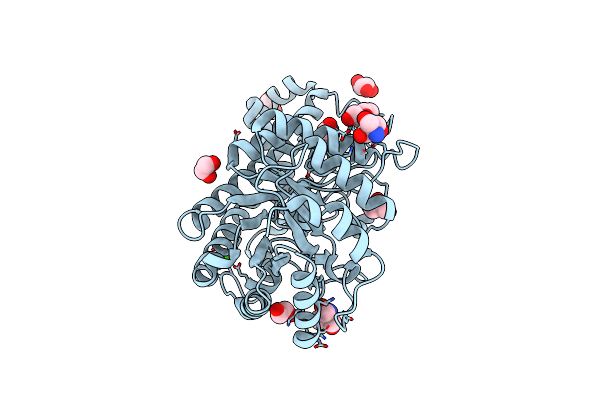 |
Crystal Structure Of The Catalytic Domain Of Beta-Amylase From Paenibacillus Polymyxa
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-02-20 Classification: HYDROLASE Ligands: CA, CL, EDO, GOL, TRS, PEG |
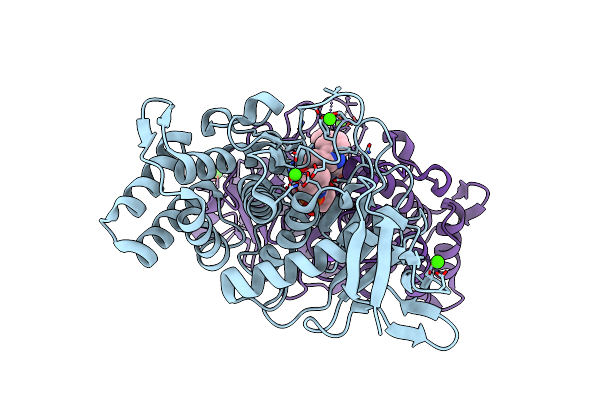 |
Crystal Structure Of Gentlyase, The Neutral Metalloprotease Of Paenibacillus Polymyxa
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2013-01-09 Classification: HYDROLASE Ligands: ZN, CA, RDF, NA |
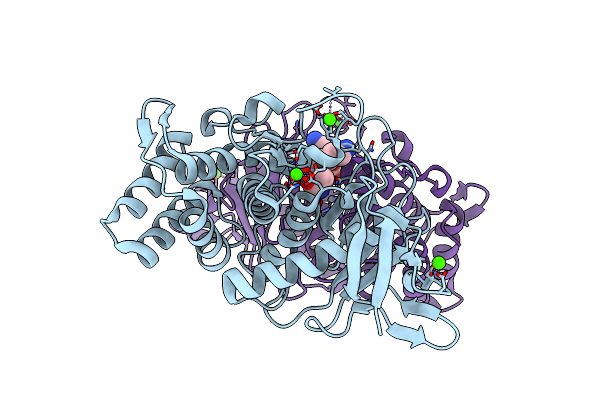 |
Crystal Structure Of Gentlyase, The Neutral Metalloprotease Of Paenibacillus Polymyxa
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2013-01-02 Classification: HYDROLASE Ligands: ZN, CA, THR, LYS |
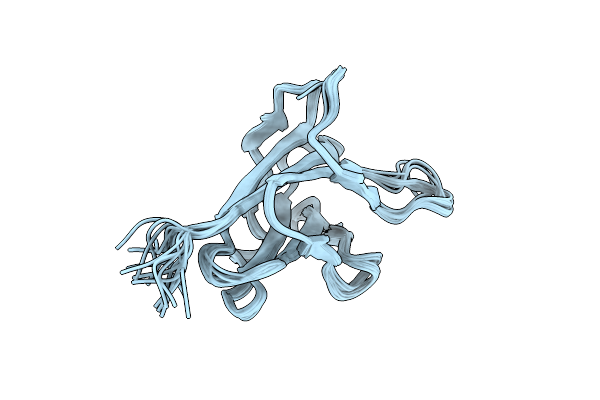 |
Solution Strucuture Of The Cbm25-1 Of Beta/Alpha-Amylase From Paenibacillus Polymyxa
Organism: Paenibacillus polymyxa
Method: SOLUTION NMR Release Date: 2012-04-04 Classification: HYDROLASE |
 |
Solution Strucuture Of The Cbm25-2 Of Beta/Alpha-Amylase From Paenibacillus Polymyxa
Organism: Paenibacillus polymyxa
Method: SOLUTION NMR Release Date: 2012-04-04 Classification: HYDROLASE |
 |
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-08-10 Classification: TOXOFLAVIN BINDING PROTEIN Ligands: MN |
 |
Crystal Structure Of Toxoflavin-Degrading Enzyme In Complex With Toxoflavin
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-08-10 Classification: TOXOFLAVIN BINDING PROTEIN Ligands: MN, TOF |
 |
Structure Of A Paenibacillus Polymyxa Xyloglucanase From Glycoside Hydrolase Family 44
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2011-06-22 Classification: HYDROLASE Ligands: BTB, CA, CL, SO4, PG4 |
 |
Structure Of A Paenibacillus Polymyxa Xyloglucanase From Glycoside Hydrolase Family 44
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2011-06-22 Classification: HYDROLASE Ligands: NOY, BGC, CA, CL, SO4, EDO |
 |
Structure Of A Paenibacillus Polymyxa Xyloglucanase From Glycoside Hydrolase Family 44
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2011-06-15 Classification: HYDROLASE Ligands: OXZ, CA, EDO, CL, SO4, PGE |
 |
Structure Of A Paenibacillus Polymyxa Xyloglucanase From Gh Family 44 With Xyloglucan
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-06-08 Classification: HYDROLASE Ligands: CA, CL, SO4, EDO |
 |
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2011-01-12 Classification: LYASE |

