Search Count: 5
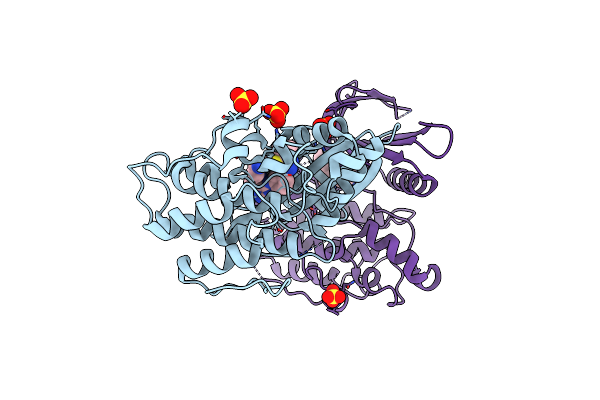 |
Crystal Structure Of Ap2 Associated Kinase 1 Complexed With 5-[(4-Aminopiperidin-1-Yl)Methyl]-N-{3-[5-(Propan-2-Yl)-1,3,4-Thiadiazol-2-Yl]Phenyl}Pyrrolo[2,1-F][1,2,4]Triazin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-07-12 Classification: TRANSFERASE Ligands: YFV, SO4 |
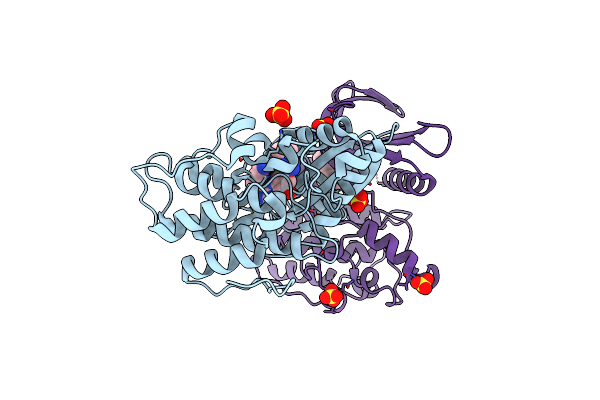 |
Crystal Structure Of Ap2 Associated Kinase 1 Complexed With (5P)-3-({(8R)-5-[(4-Aminopiperidin-1-Yl)Methyl]Pyrrolo[2,1-F][1,2,4]Triazin-4-Yl}Amino)-5-[2-(Propan-2-Yl)-2H-Tetrazol-5-Yl]Phenol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-07-12 Classification: TRANSFERASE Ligands: ZRR, SO4 |
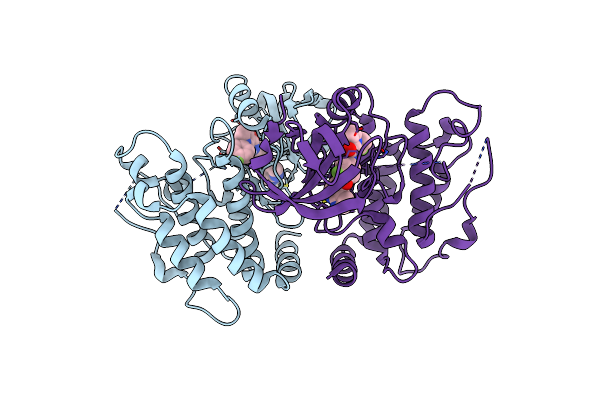 |
Crystal Structure Of Mripk3 Complexed With N-(3-Fluoro-4-{1H-Pyrrolo[2,3-B]Pyridin-4-Yloxy}Phenyl)-1-(4-Fluorophenyl)-2-Oxo-1,2-Dihydropyridine-3-Carboxamide
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-07-17 Classification: TRANSFERASE Ligands: 1FN |
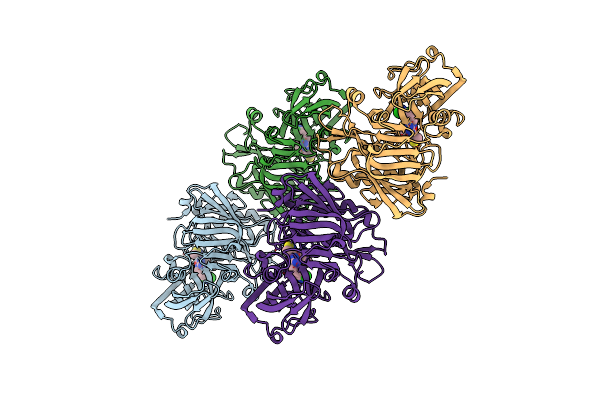 |
Crystal Structure Of Beta-Site App-Cleaving Enzyme 1 (Bace-Wt) Complex With N-(N-(4-Amino-3,5- Dichlorobenzyl)Carbamimidoyl)-3-(4-Methoxyphenyl)-5- Methyl-4-Isothiazolecarboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2012-10-10 Classification: HYDROLASE/INHIBITOR Ligands: 0VA, IOD |
 |
Crystal Structure Of Beta-Site App-Cleaving Enzyme 1 (Bace-Db-Mut) Complex With N-(N-(4- Acetamido-3-Chloro-5-Methylbenzyl)Carbamimidoyl)-3-(4- Methoxyphenyl)-5-Methyl-4-Isothiazolecarboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-10-10 Classification: HYDROLASE/INHIBITOR Ligands: 0VB, IOD |

