Search Count: 602
 |
Structure Of The Transaminase Phnw From Vibrio Vulnificus In Complex With Plp
Organism: Vibrio vulnificus nbrc 15645 = atcc 27562
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSFERASE |
 |
Structure Of The Transaminase Phnw From Vibrio Vulnificus In Complex With Plp And Aep
Organism: Vibrio vulnificus nbrc 15645 = atcc 27562
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSFERASE Ligands: P7I |
 |
Structure Of The K193M Mutant Of Transaminase Phnw From Vibrio Vulnificus In Complex With Plp And Aep
Organism: Vibrio vulnificus nbrc 15645 = atcc 27562
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSFERASE Ligands: PLP, P7I, CL |
 |
Organism: Caldanaerobacter subterraneus subsp. tengcongensis
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: ISOMERASE Ligands: B12, 5AD |
 |
Organism: Streptomyces lincolnensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-08-14 Classification: LYASE Ligands: GOL |
 |
Closed State Of Lysine 5,6-Aminomutase From Thermoanaerobacter Tengcongensis
Organism: Caldanaerobacter subterraneus subsp. tengcongensis
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: ISOMERASE Ligands: B12, 5AD, X6I |
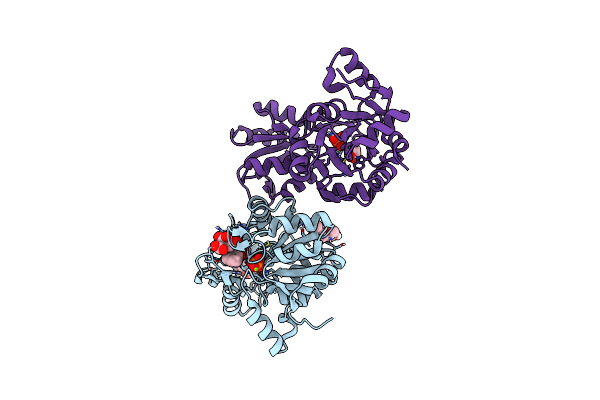 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-06-12 Classification: HYDROLASE Ligands: MG, PO4, UK9, PGE, CIT, GOL |
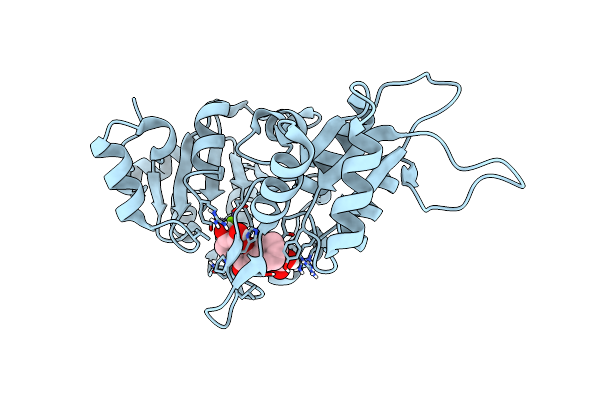 |
Human Pyridoxal Phosphatase In Complex With 7,8-Dihydroxyflavone Without Phosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-06-12 Classification: HYDROLASE Ligands: UK9, FMT, MG, CL |
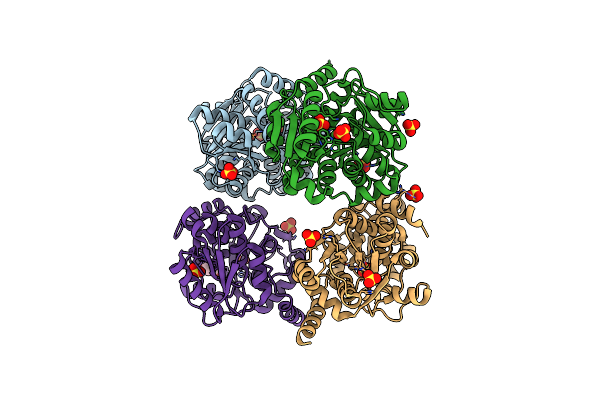 |
Crystal Structure Of Mutant Catalytic Domains Of Threonine Deaminase In Complex With Plp
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2024-06-12 Classification: STRUCTURAL PROTEIN Ligands: SO4, GOL |
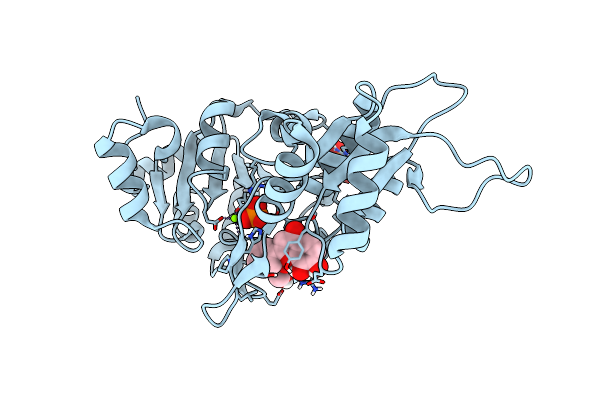 |
Human Pyridoxal Phosphatase In Complex With 7,8-Dihydroxyflavone And Phosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-06-12 Classification: HYDROLASE Ligands: MG, UK9, GOL, PO4 |
 |
Organism: Streptomyces albogriseolus 1-36
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2024-05-01 Classification: TRANSFERASE Ligands: PLP, PLR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-02-07 Classification: OXIDOREDUCTASE Ligands: BME, FMN, PLP, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2024-02-07 Classification: OXIDOREDUCTASE Ligands: PO4, FMN |
 |
Promiscuous Amino Acid Gamma Synthase From Caldicellulosiruptor Hydrothermalis In Open Conformation
Organism: Caldicellulosiruptor hydrothermalis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-12-27 Classification: TRANSFERASE |
 |
Promiscuous Amino Acid Gamma Synthase From Caldicellulosiruptor Hydrothermalis In Closed Conformation
Organism: Caldicellulosiruptor hydrothermalis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-12-27 Classification: TRANSFERASE Ligands: MES, NA, AE3, ETE, 1PE |
 |
Human Holo Aromatic L-Amino Acid Decarboxylase (Aadc) Native Structure At Physiological Ph
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-08-30 Classification: LYASE Ligands: PLP, PG4 |
 |
Human Holo Aromatic L-Amino Acid Decarboxylase (Aadc) External Aldimine With L-Dopa Methylester
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-08-30 Classification: LYASE Ligands: PLP, W5I, PG4 |
 |
Structure Of Gntc, A Plp-Dependent Enzyme Catalyzing L-Enduracididine Biosynthesis From (S)-4-Hydroxy-L-Arginine
Organism: Dolichospermum flos-aquae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-04-12 Classification: BIOSYNTHETIC PROTEIN Ligands: MG |
 |
Structure Of Gntc, A Plp-Dependent Enzyme Catalyzing L-Enduracididine Biosynthesis From (S)-4-Hydroxy-L-Arginine, With The Substrate Bound
Organism: Dolichospermum flos-aquae
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2023-04-12 Classification: BIOSYNTHETIC PROTEIN Ligands: MG, XUR |
 |
1.55 A X-Ray Crystallographic Structure Of Saph From Streptomyces Sp. (Hph0547) Involved In Pseudouridimycin Biosynthesis
Organism: Streptomyces sp. hph0547
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-02-15 Classification: BIOSYNTHETIC PROTEIN Ligands: PMP, GOL, K |

