Search Count: 213
 |
Organism: Raoultella planticola
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: LYASE Ligands: ADP, MG, PYR, MES, CL |
 |
Crystal Structure Of Dihydrodipicolinate Synthase From Mycobacterium Tuberculosis In Complex With Pyruvate
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: LYASE Ligands: PYR, GOL, MG, EDO, CL, PEG |
 |
Crystal Structure Of The Coxiella Burnetii 2-Methylisocitrate Lyase Bound To Products Succinic And Pyruvic Acid
Organism: Coxiella burnetii
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: CYTOSOLIC PROTEIN Ligands: SIN, PYR, EDO, MG, CL, DMS, PGE, PEG |
 |
Crystal Structure Of Trans-O-Hydroxybenzylidenepyruvate Hydratase-Aldolase From Pseudomonas Fluorescens N3 Bound To Pyruvate
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: LYASE Ligands: PYR, PO4, GOL |
 |
Organism: Homo sapiens, Mus musculus, Synthetic construct, Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:2.78 Å Release Date: 2025-03-05 Classification: TRANSPORT PROTEIN/IMMUNE SYSTEM Ligands: PYR |
 |
Crystal Structure Of The G200R Mutant From The Maize Chloroplastic Photosynthetic Nadp(+)-Dependent Malic Enzyme
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-01-15 Classification: PHOTOSYNTHESIS Ligands: PYR |
 |
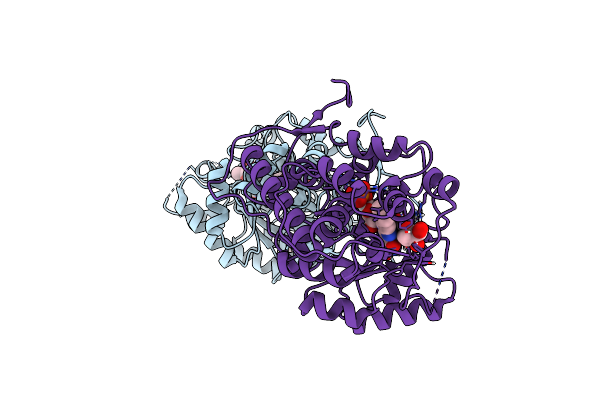
Crystal Structure Of Rida Family Protein Pa5083 From Pseudomonas Aeruginosa With Pyruvic Acid Bound
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2024-09-04 Classification: HYDROLASE Ligands: PYR, SO4 |
 |
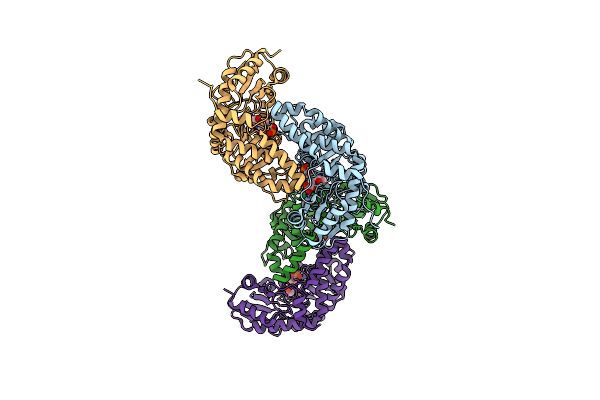
Organism: Aerococcus viridans
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2024-08-14 Classification: FLAVOPROTEIN Ligands: FMN, EDO, LAC, PYR |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-03-06 Classification: LYASE Ligands: PLP, PYR |
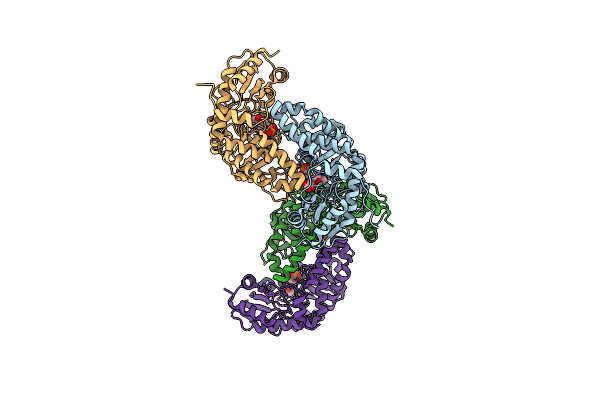 |
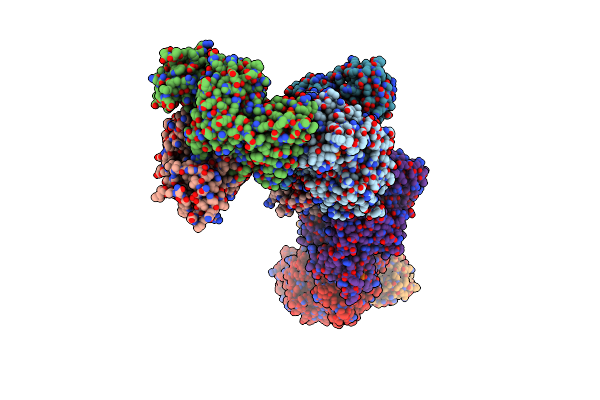
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-03-06 Classification: LYASE Ligands: PLP, PYR |
 |
Structure Of Human Erythrocyte Pyruvate Kinase In Complex With An Allosteric Activator Ag-946
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-12-27 Classification: TRANSFERASE/ACTIVATOR Ligands: FBP, MN, K, PYR, HVI |
 |
Structure Of Human Erythrocyte Pyruvate Kinase In Complex With An Allosteric Activator Compound 2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2023-12-27 Classification: TRANSFERASE/ACTIVATOR Ligands: FBP, PYR, MN, K, I07 |
 |
Structure Of Human Erythrocyte Pyruvate Kinase In Complex With An Allosteric Activator Compound 12
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-12-27 Classification: TRANSFERASE/ACTIVATOR Ligands: I0R, PYR, FBP, MN, K |
 |
Crystal Structure Of Macrophage Migration Inhibitory Factor (Mif) From Trichomonas Vaginalis (I41 Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-08 Classification: CYTOKINE Ligands: IOD, PYR |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2023-10-18 Classification: OXIDOREDUCTASE Ligands: FAD, MN, PYR |
 |
Selenomethionine Derivatized Citrate Synthase (Cita) In Mycobacterium Tuberculosis With Pyruvate
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-06-07 Classification: CYTOSOLIC PROTEIN Ligands: EDO, PYR |
 |
Pyruvate Bound Structure Of Citrate Synthase (Cita) In Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2023-06-07 Classification: CYTOSOLIC PROTEIN Ligands: PYR, PGE, EDO, PEG |
 |
Rabbit Muscle Pyruvate Kinase In Complex With Magnesium, Potassium And Pyruvate
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-05-24 Classification: CYTOSOLIC PROTEIN Ligands: GOL, EDO, K, MG, PYR, TRS |
 |
Organism: Geobacillus kaustophilus
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-04-05 Classification: OXIDOREDUCTASE Ligands: NAD, EDO, PYR |
 |
Crystal Structure Of L-2,4-Diketo-3-Deoxyrhamnonate Hydrolase From Sphingomonas Sp. (Pyruvate Bound-Form)
Organism: Sphingomonas sp. ska58
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2023-02-08 Classification: HYDROLASE Ligands: MG, PYR |

