Search Count: 13
 |
Proline Racemase (Pror) From The Gram-Positive Bacterium Acetoanaerobium Sticklandii From Isotropic Tetragonal Data At 3.15 A
Organism: Acetoanaerobium sticklandii dsm 519
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2023-07-05 Classification: ISOMERASE Ligands: PYC |
 |
Proline Racemase (Pror) From The Gram-Positive Bacterium Acetoanaerobium Sticklandii From Isotropic Orthorhombic Data At 3.59 A
Organism: Acetoanaerobium sticklandii dsm 519
Method: X-RAY DIFFRACTION Resolution:3.59 Å Release Date: 2023-07-05 Classification: ISOMERASE Ligands: PYC |
 |
Structural And Functional Analysis Of The Proline Racemase (Pror) From The Gram-Positive Bacterium Acetoanaerobium Sticklandii
Organism: Acetoanaerobium sticklandii (strain atcc 12662 / dsm 519 / jcm 1433 / ncimb 10654)
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2022-08-10 Classification: ISOMERASE Ligands: PYC |
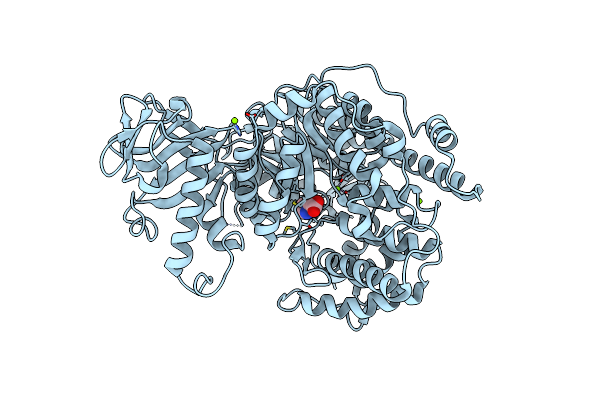 |
Crystal Structure Of Mycobacterium Tuberculosis Malate Synthase In Complex With 1H-Pyrrole-2-Carboxylic Acid
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-08-10 Classification: TRANSFERASE Ligands: MG, PYC |
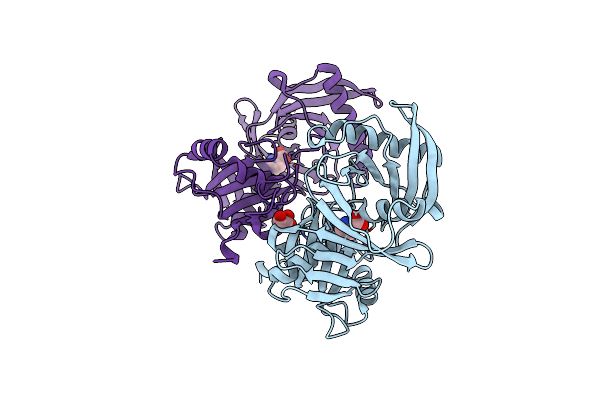 |
Crystal Structure Of A 4-Hydroxyproline Epimerase From Burkholderia Multivorans Atcc 17616, Target Efi-506586, Open Form, With Bound Pyrrole-2-Carboxylate
Organism: Burkholderia multivorans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-05-07 Classification: ISOMERASE Ligands: GOL, PYC |
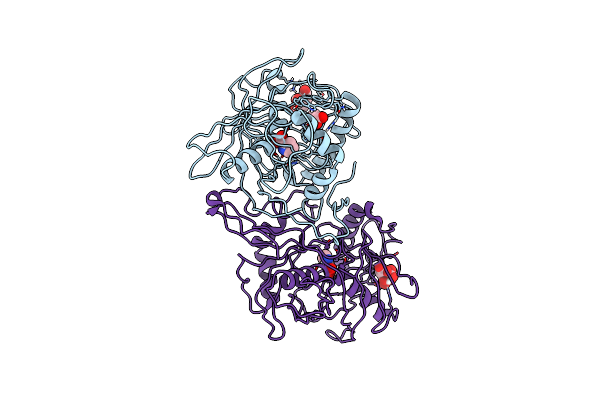 |
Crystal Structure Of A 3-Hydroxyproline Dehydratse From Agrobacterium Vitis, Target Efi-506470, With Bound Pyrrole 2-Carboxylate, Ordered Active Site
Organism: Agrobacterium vitis s4
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-05-01 Classification: ISOMERASE Ligands: CIT, ACT, PYC |
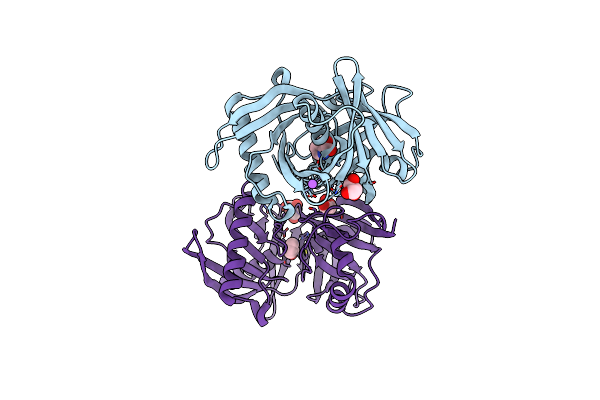 |
Crystal Structure Of The Complex Of A Hydroxyproline Epimerase (Target Efi-506499, Pseudomonas Fluorescens Pf-5) With The Inhibitor Pyrrole-2-Carboxylate
Organism: Pseudomonas protegens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-03-13 Classification: Isomerase/Isomerase Inhibitor complex Ligands: GOL, PYC, NA |
 |
Heterotetrameric Sarcosine Oxidase From Corynebacterium Sp. U-96 In Complex With Pyrrole 2-Carboxylate
Organism: Corynebacterium sp. u-96
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-08-25 Classification: OXIDOREDUCTASE Ligands: NAD, SO4, FAD, FMN, PYC, ZN |
 |
Proline Racemase In Complex With 2 Molecules Of Pyrrole-2-Carboxylic Acid (Holo Form)
Organism: Trypanosoma cruzi
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-02-03 Classification: RACEMASE Ligands: PYC |
 |
Proline Racemase In Complex With One Molecule Of Pyrrole-2-Carboxylic Acid (Hemi Form)
Organism: Trypanosoma cruzi (strain cl brener)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2006-02-03 Classification: RACEMASE Ligands: PYC |
 |
Crystal Structure Of Delta1-Piperideine-2-Carboxylate Reductase From Pseudomonas Syringae Complexed With Nadph And Pyrrole-2-Carboxylate
Organism: Pseudomonas syringae pv. tomato
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2005-10-04 Classification: OXIDOREDUCTASE Ligands: NDP, PYC |
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2002-08-30 Classification: OXIDOREDUCTASE Ligands: CL, FAD, PYC |
 |
Complex Of Monomeric Sarcosine Oxidase With The Inhibitor Pyrrole-2-Carboxylate
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-12-06 Classification: OXIDOREDUCTASE Ligands: PO4, CL, FAD, PYC |

