Search Count: 86
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-01-08 Classification: MEMBRANE PROTEIN Ligands: MPD, PX4 |
 |
Homomeric Alpha3 Glycine Receptor In The Presence Of 1 Mm Glycine At Ph 6.4 In A Desensitized State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: MEMBRANE PROTEIN Ligands: GLY, PIO, PX4 |
 |
Homomeric Alpha3 Glycine Receptor In The Presence Of 0.1 Mm Glycine In A Desensitized State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: MEMBRANE PROTEIN Ligands: GLY, PIO, PX4 |
 |
Homomeric Alpha3 Glycine Receptor In The Presence Of 0.1 Mm Glycine In An Apo State.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: MEMBRANE PROTEIN Ligands: PIO, PX4, CLR |
 |
Homomeric Alpha3 Glycine Receptor In The Presence Of 1 Mm Glycine In An Desensitized State.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: MEMBRANE PROTEIN Ligands: GLY, PIO, PX4 |
 |
Homomeric Alpha3 Glycine Receptor In The Presence Of 0.1 Mm Glycine At Ph 6.4 In A Desensitized State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: MEMBRANE PROTEIN Ligands: GLY, PIO, PX4 |
 |
Homomeric Alpha3 Glycine Receptor In The Presence Of 0.1 Mm Glycine At Ph 6.4 In An Apo State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: MEMBRANE PROTEIN Ligands: PX4, PIO, GLY |
 |
Homomeric Alpha3 Glycine Receptor In The Presence Of 0.1 Mm Glycine At Ph 6.4 In An Intermediate State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: MEMBRANE PROTEIN Ligands: PIO, PX4, GLY |
 |
Homomeric Alpha3 Glycine Receptor In The Presence Of 0.1 Mm Glycine And 0.1 Mm Zinc In A Desensitized State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: MEMBRANE PROTEIN Ligands: GLY, ZN, PIO, PX4 |
 |
Homomeric Alpha3 Glycine Receptor In The Presence Of 0.1 Mm Glycine And 0.1 Mm Zinc In An Apo State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: MEMBRANE PROTEIN Ligands: ZN, PX4 |
 |
Homomeric Alpha3 Glycine Receptor In The Presence Of 0.1 Millimolar Glycine And 1 Micromolar Zinc In A Desensitized State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: MEMBRANE PROTEIN Ligands: GLY, ZN, PIO, PX4 |
 |
Crystal Structure Of Navab I22V As A Basis For The Human Nav1.7 Inherited Erythromelalgia I136V Mutation
Organism: Aliarcobacter butzleri rm4018
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2023-10-25 Classification: MEMBRANE PROTEIN Ligands: BGC, PX4, CPS |
 |
Crystal Structure Of Navab E96P As A Basis For The Human Nav1.7 Inherited Erythromelalgia S211P Mutation
Organism: Aliarcobacter butzleri rm4018
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2023-10-25 Classification: MEMBRANE PROTEIN Ligands: PX4, CPS |
 |
Structure Of Navab L98R As A Basis For The Human Nav1.7 Inherited Erythromelalgia L823R Mutation
Organism: Aliarcobacter butzleri rm4018
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2023-10-25 Classification: MEMBRANE PROTEIN Ligands: PX4 |
 |
Crystal Structure Of Navab L101S As A Basis For The Human Nav1.7 Inherited Erythromelalgia F216S Mutation
Organism: Aliarcobacter butzleri rm4018
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2023-10-25 Classification: MEMBRANE PROTEIN Ligands: PX4, CPS |
 |
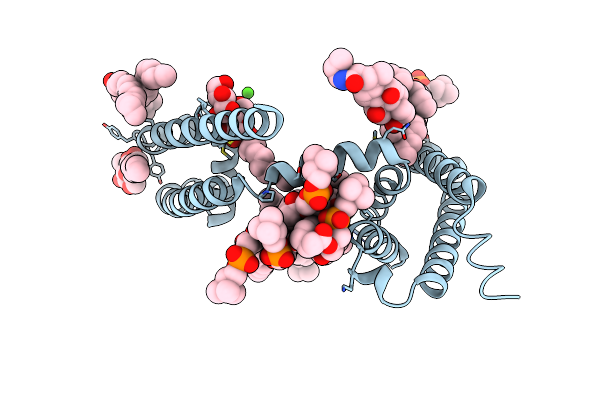
Crystal Structure Of Voltage-Gated Sodium Channel Navab N49K Mutant In Sodium Ion Condition
Organism: Aliarcobacter butzleri
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2023-07-26 Classification: MEMBRANE PROTEIN Ligands: LMT, 1N7, PX4, NA |
 |
Crystal Structure Of Voltage-Gated Sodium Channel Navab N49K Mutant In Calcium Ion Condition
Organism: Aliarcobacter butzleri
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-07-26 Classification: MEMBRANE PROTEIN Ligands: LMT, 1N7, PX4, CA |
 |
Crystal Structure Of Voltage-Gated Sodium Channel Navab N49K/L176Q Mutant In Sodium Ion Condition
Organism: Aliarcobacter butzleri
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2023-07-26 Classification: MEMBRANE PROTEIN Ligands: LMT, 1N7, PX4, NA |
 |
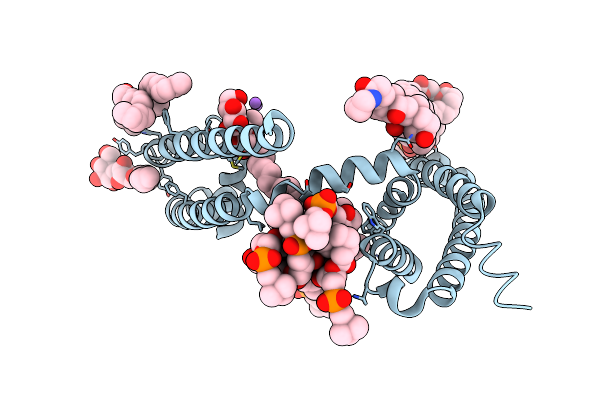
Crystal Structure Of Voltage-Gated Sodium Channel Navab N49K/L176Q Mutant In Calcium Ion Condition
Organism: Aliarcobacter butzleri
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2023-07-26 Classification: MEMBRANE PROTEIN Ligands: LMT, 1N7, PX4, CA |
 |
Crystal Structure Of Voltage-Gated Sodium Channel Navab N49K/L176G Mutant In Sodium Ion Condition
Organism: Aliarcobacter butzleri
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2023-07-26 Classification: MEMBRANE PROTEIN Ligands: LMT, 1N7, PX4, NA |

