Search Count: 1,521
All
Selected
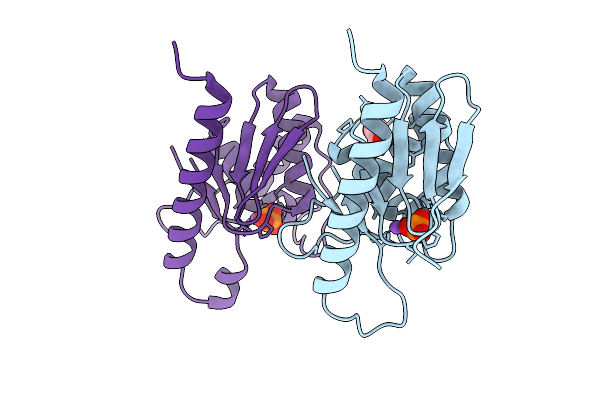 |
High Resolution Structure Of Cytidine Deaminase T6S Toxin From Pseudomonas Syringae
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2025-11-12 Classification: TOXIN Ligands: ZN, PO4, EDO, NA |
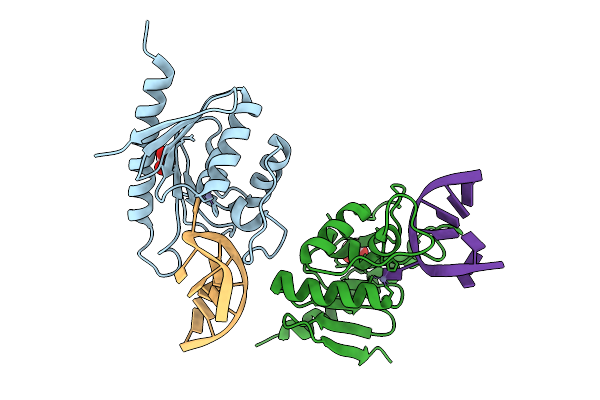 |
Cytidine Deaminase T6S Toxin From Pseudomonas Syringae Complexed With Substrate Dna
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-11-12 Classification: TOXIN/DNA Ligands: ZN, EDO |
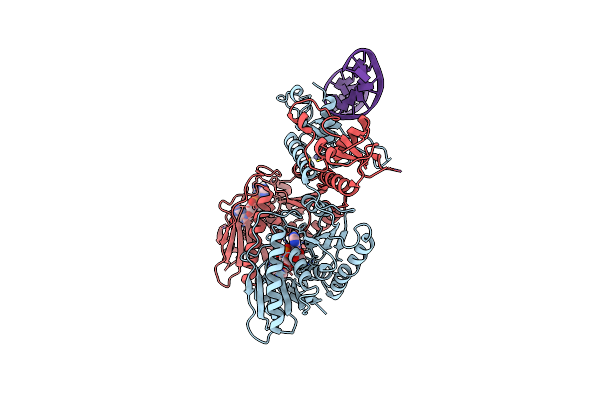 |
Cbass Pseudomonas Syringae Cap5 Tetramer With Dna Duplex And 3'2'-C-Gamp Cyclic Dinucleotide Ligand
Organism: Pseudomonas syringae, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2025-06-18 Classification: IMMUNE SYSTEM Ligands: 4UR, ZN |
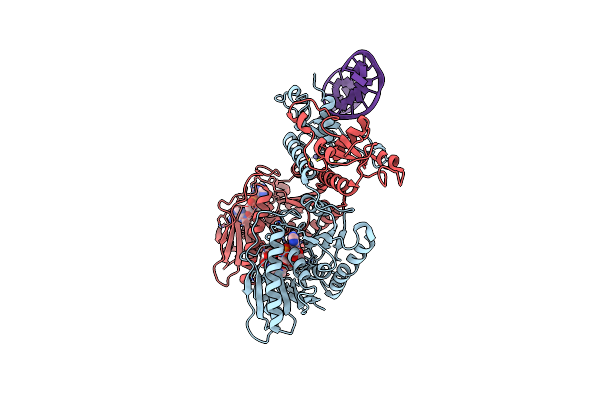 |
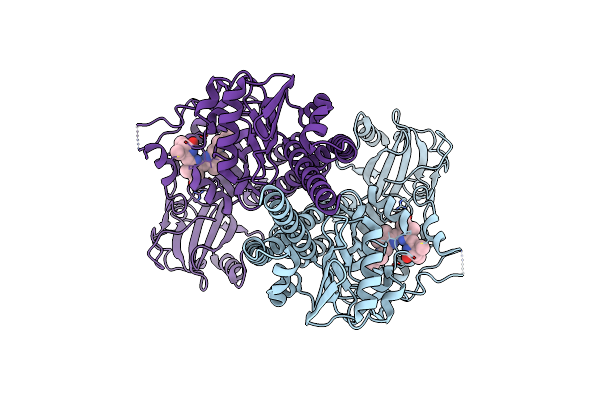
Cbass Pseudomonas Syringae Cap5 Tetramer With Dna Duplex And 3'2'-C-Diamp Cyclic Dinucleotide Ligand
Organism: Pseudomonas syringae, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-06-18 Classification: IMMUNE SYSTEM Ligands: ZN, Y4F, GOL |
 |
Cbass Pseudomonas Syringae Cap5 Tetramer With 3'2'-C-Gamp Cyclic Dinucleotide Ligand (His56Ala Mutant Without Mg2+ Ions)
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-06-18 Classification: IMMUNE SYSTEM Ligands: ZN, 4UR, GOL |
 |
Crystal Structure Of P. Syringae Phosphinothricin Acetyltransferase Pspto_3321
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: CIT, PEG, NA, PGE |
 |
Crystal Structure Of P. Syringae Phosphinothricin Acetyltransferase Pspto_3321 In Complex With L-Phosphinothricin
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: SO4, PPQ, ACT, NA, EDO, PEG |
 |
Crystal Structure Of Levansucrase From Pseudomonas Syringae In Complex With A Tetravalent Iminosugar
Organism: Pseudomonas syringae pv. actinidiae
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-10-02 Classification: TRANSFERASE Ligands: MES, GOL, VXT, 15P, DMS |
 |
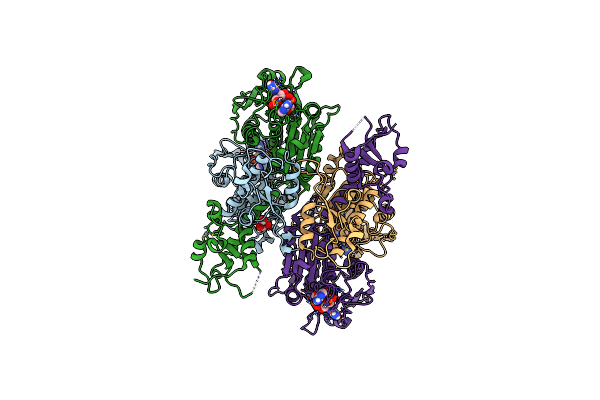
Crystal Structure Of The Levansucrase Beta From Pseudomonas Syringae Pv. Actinidiae
Organism: Pseudomonas syringae pv. actinidiae
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2024-09-25 Classification: TRANSFERASE Ligands: PO4, P4K, ACT, GOL, SCN, MES |
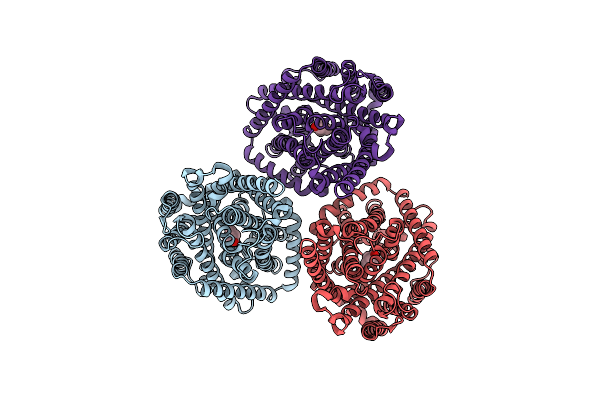 |
Organism: Pseudomonas syringae
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: CHOLINE-BINDING PROTEIN Ligands: CHT |
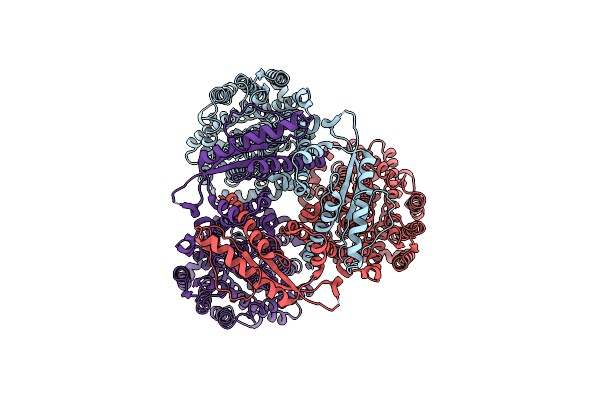 |
Organism: Pseudomonas syringae
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: CHOLINE-BINDING PROTEIN |
 |
Crystal Structure Of Endoribonuclease L-Psp Family Protein Pspto0102 Sulfur Sad Phased
Organism: Pseudomonas syringae pv. tomato
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2024-09-04 Classification: HYDROLASE |
 |
Organism: Pseudomonas syringae pv. tomato
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-09-04 Classification: HYDROLASE |
 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: PLANT PROTEIN Ligands: LBV |
 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: PLANT PROTEIN Ligands: LBV |
 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: PLANT PROTEIN Ligands: LBV |
 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: PLANT PROTEIN Ligands: LBV |
 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: PLANT PROTEIN Ligands: LBV |
 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: PLANT PROTEIN Ligands: LBV |
 |
Crystal Structure Of Alpha Keto Acid C-Methyl-Transferases Mrsa Native-Form
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: MG |

