Search Count: 3,374
All
Selected
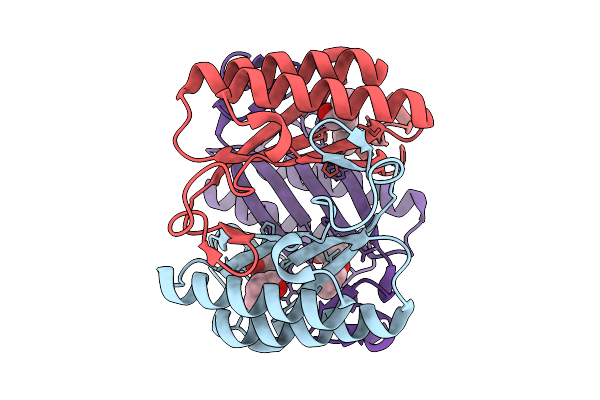 |
X-Ray Crystallographic Structure Of 4-Ot (F11) In Complex With 2-Hydroxycinnamaldehyde
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2026-01-21 Classification: LYASE Ligands: A1IZH |
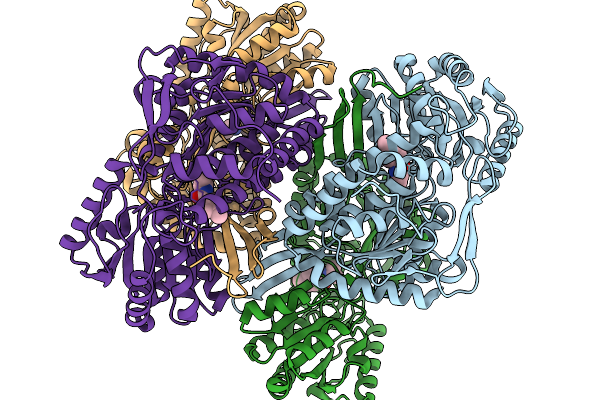 |
Co-Crystal Structure Of Phbdd-L163G With Product N-Isopropyl-4-Methylbenzamide
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: A1EBJ |
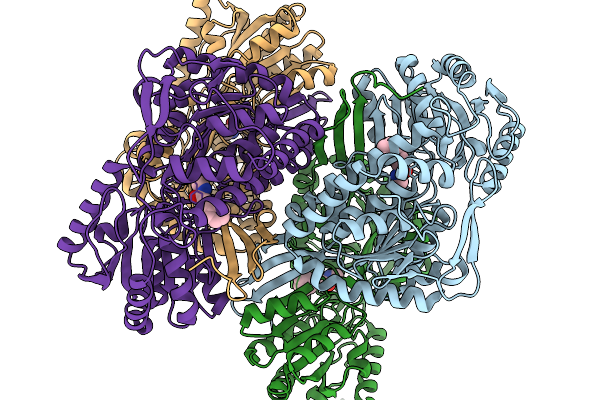 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: A1EBQ |
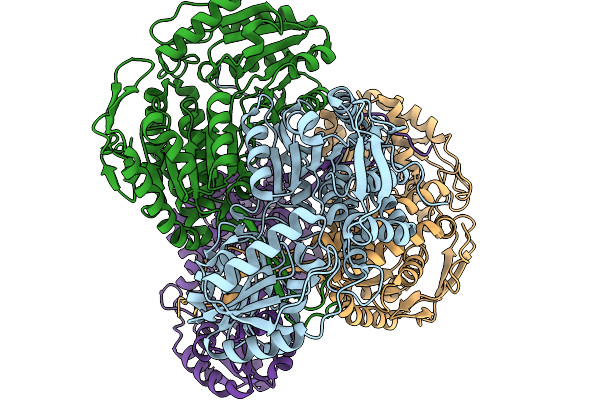 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2026-01-14 Classification: OXIDOREDUCTASE |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: A1EC9 |
 |
Co-Crystal Structure Of Phbdd-M1-L163G-T234A With N-Cyclopropyl-4-Methylbenzamide
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: A1EDB |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: A1EDA, SO4 |
 |
Co-Crystal Structure Of Phbdd-M1 With 1H-Pyrrolo[2,3-B]Pyridine-4-Carbaldehyde
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: A1EDC |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2026-01-14 Classification: OXIDOREDUCTASE |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-01-14 Classification: ISOMERASE Ligands: A1CF8, EDO, MG |
 |
Pseudomonas Putida Pore-Forming Toxin Tke5 In Complex With Its Cognate Type Vi Adaptor Protein Tap3
Organism: Pseudomonas putida kt2440
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: TOXIN |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2026-01-07 Classification: OXIDOREDUCTASE Ligands: HEM, ACT |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2025-11-12 Classification: DNA BINDING PROTEIN Ligands: ACY, SO4, HCI |
 |
Crystal Structure Of The Pseudomonas Putida Xre-Res Toxin-Antitoxin Complex Bound To Promoter Dna
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-08-20 Classification: GENE REGULATION |
 |
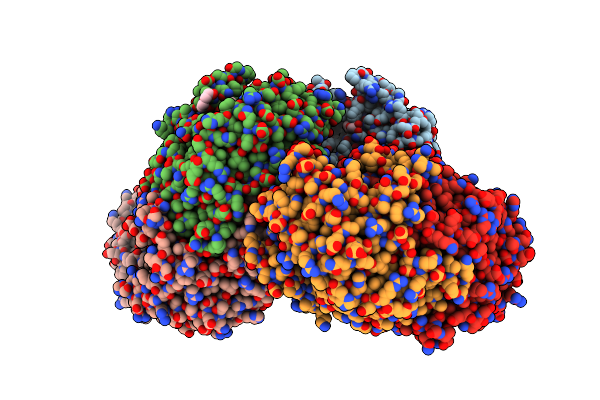
Crystal Structure Of Omega Transaminase Ta_2799 From Pseudomonas Putida Kt2440
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: GOL, EDO, PEG |
 |
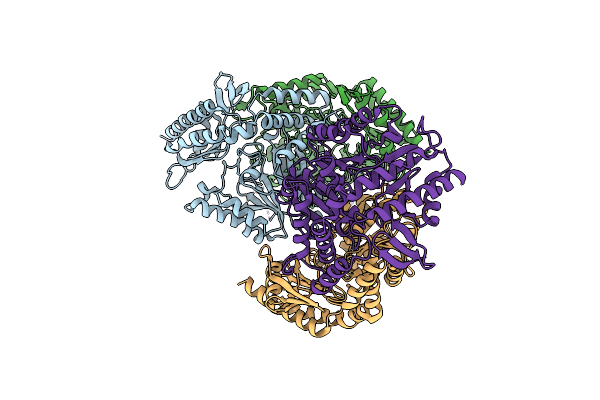
Crystal Structure Of The L322F Mutant Of Omega Transaminase Ta_2799 From Pseudomonas Putida Kt2440
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: PEG, EDO, SO4, GOL |
 |
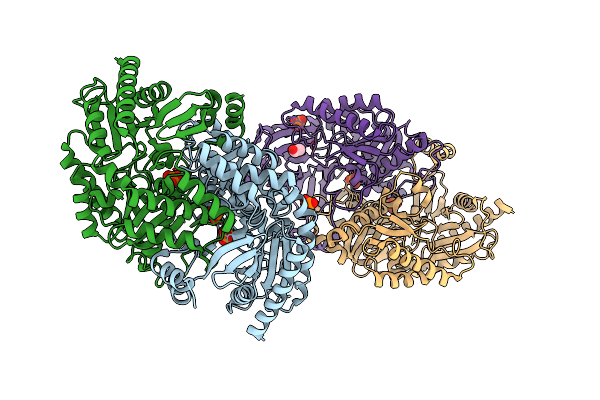
Crystal Structure Of The Open State Of Omega Transaminase Ta_5182 From Pseudomonas Putida Kt2440
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: SO4 |
 |
Crystal Structure Of The Closed State Of The Omega Transaminase Ta_5182 From Pseudomonas Putida Kt2440
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: PO4, EDO |
 |
Crystal Structure Of The Dyp-Type Peroxidase Pross Variant From Pseudomonas Putida
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: OXY, HEM, CL |
 |
Crystal Structure Of A Dyp-Type Peroxidase Fireprot Variant From Pseudomonas Putida
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: GOL, HEM, CL |

