Search Count: 75
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.52 Å Release Date: 2025-01-22 Classification: ELECTRON TRANSPORT Ligands: LOP, CDL, HEM, A1IKG, LMT, HEC, FES, PSC, SO4 |
 |
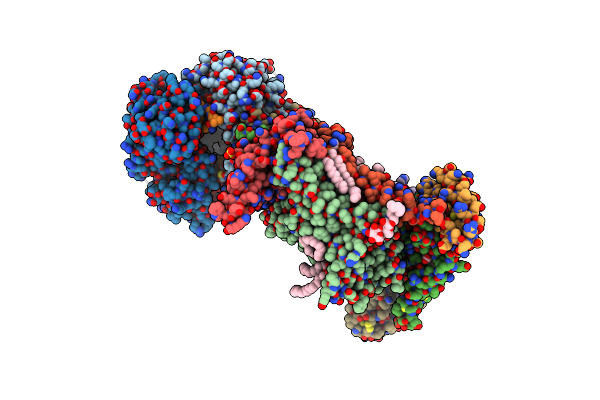
In-Situ Structure Of Typea Supercomplex With Lipids In Respiratory Chain (Composite)
|
 |
High Resolution In-Situ Structure Of Typea Supercomplex In Respiratory Chain (I1Iii2Iv1,Composite)
|
 |
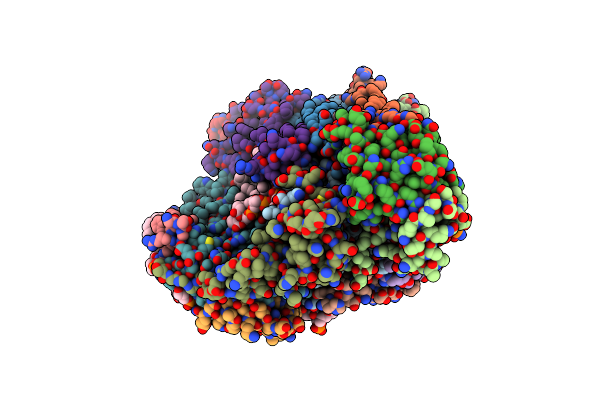 |
High Resolution In-Situ Structure Of Complex Iv In Respiratory Supercomplex
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: ELECTRON TRANSPORT Ligands: PGV, HEA, CU, MG, NA, CDL, CUA, PSC, ZN, PEK, PO4 |
 |
 |
 |
Organism: Bombyx mori
Method: ELECTRON MICROSCOPY Release Date: 2024-01-10 Classification: MEMBRANE PROTEIN Ligands: PSC, PC7, 9Z9 |
 |
Cryoem Structure Of Insect Gustatory Receptor Bmgr9 In The Presence Of Fructose
Organism: Bombyx mori
Method: ELECTRON MICROSCOPY Release Date: 2024-01-10 Classification: MEMBRANE PROTEIN Ligands: PSC |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2023-09-27 Classification: MEMBRANE PROTEIN Ligands: CU, MG, NA, PGV, HEA, OH, CUA, TGL, PSC, CHD, PEK, CDL, DMU, ZN, SAC |
 |
Time-Resolve Sfx Structure Of A Photoproduct Of Carbon Monoxide Complex Of Bovine Cytochrome C Oxidase
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-09-20 Classification: MEMBRANE PROTEIN Ligands: HEA, CU, MG, NA, TGL, PGV, CMO, CUA, CHD, OH, CDL, PSC, ZN, PEK, DMU, SAC |
 |
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr1 In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.56 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr2 In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.53 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr1 H225F Mutant In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.66 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
Bovine Heart Cytochrome C Oxidase In The Nitric Oxide-Bound Fully Reduced State At 100 K
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-04-12 Classification: OXIDOREDUCTASE Ligands: HEA, NO, CU, MG, NA, TGL, PGV, EDO, CUA, CHD, PEK, CDL, DMU, PSC, ZN |
 |
Crystal Structure Of Bovine Heart Cytochrome C Oxidase, Apo Structure With Dmso
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-12-21 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PER, PGV, TGL, CUA, CDL, CHD, PEK, PSC, ZN, DMU |
 |
Crystal Structure Of Bovine Heart Cytochrome C Oxidase, The Structure Complexed With An Allosteric Inhibitor T113
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-12-21 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PER, TGL, J6X, CUA, CHD, PSC, PEK, PGV, CDL, DMU, ZN |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2022-12-07 Classification: MEMBRANE PROTEIN Ligands: HEA, CU, MG, NA, CYN, TGL, PGV, EDO, CUA, CHD, PEK, CDL, DMU, PSC, ZN, PO4 |
 |
Bovine Heart Cytochrome C Oxidase In The Cyanide-Bound Fully Oxidized State At 50 K
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-11-16 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, CYN, PGV, EDO, TGL, CUA, PSC, CHD, PEK, CDL, DMU, ZN, PO4 |


