Search Count: 325
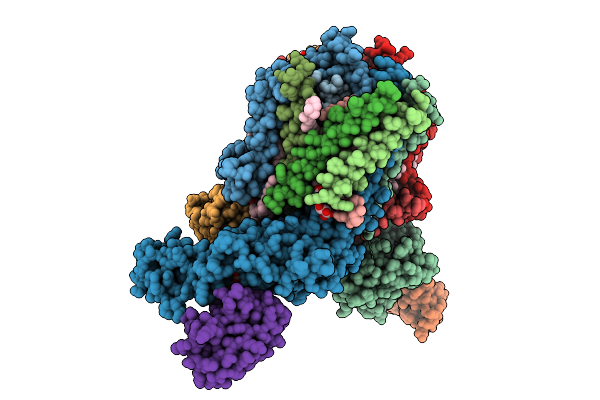 |
Cryo-Em Structure Of Spinacia Oleracea Cytochrome B6F Complex With Bound Plastocyanin
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: HEM, HEC, UMQ, PL9, CLA, FES, SQD, BCR, CU |
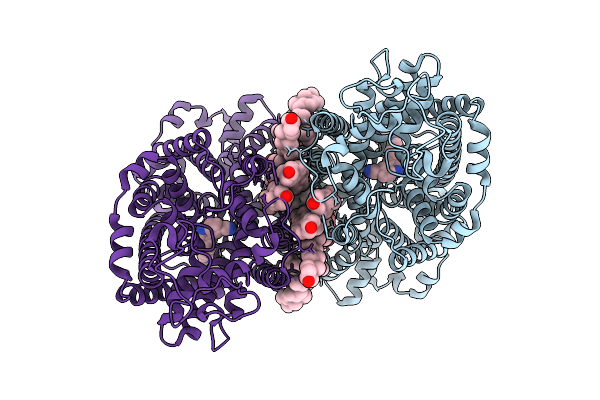 |
Cryo-Em Structure Of Lipid-Mediated Dimer Of Human Norepinephrine Transporter Net In The Presence Of The Antidepressant Vilazodone In An Inward-Open State At Resolution Of 2.44 Angstrom.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: PROTON TRANSPORT Ligands: YG7, CLR, A1EFM |
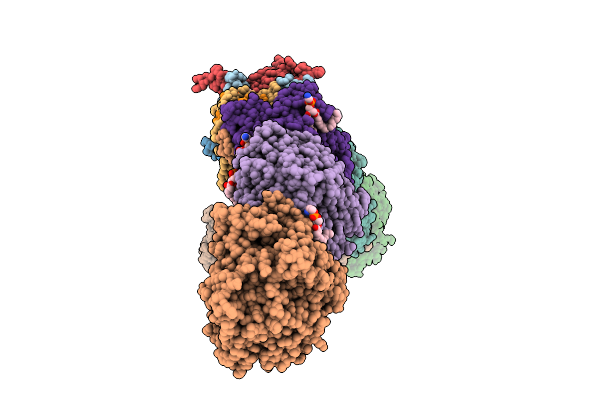 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: 3PE, SF4, FES, FMN, CA, CL |
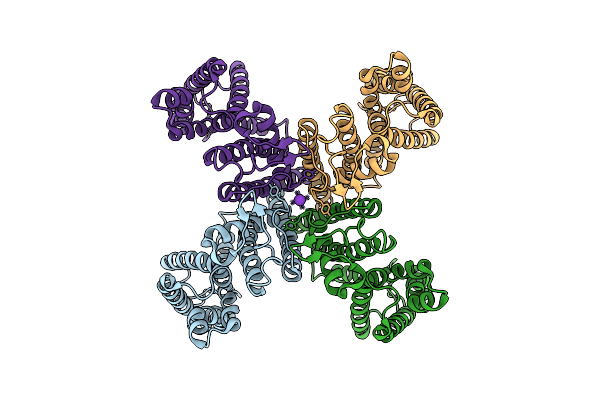 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PLANT PROTEIN Ligands: K |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PROTON TRANSPORT Ligands: K |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PROTON TRANSPORT Ligands: K |
 |
Cryo-Em Structure Of The Light-Driven Proton Pump Pspr In Detergent Micelle
Organism: Candidatus pseudothioglobus sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA, RET, LMT |
 |
Cryo-Em Structure Of The Double Mutant H84V/E120G Of The Flotillin-Associated Rhodopsin Psfar In Detergent Micelle
Organism: Candidatus pseudothioglobus sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Cryo-Em Structure Of Inner Membrane Tolqra Complex In Cymal-6-Neopentyl Glycol Detergent Micelles
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: PROTON TRANSPORT |
 |
Cryo-Em Structure Of Inner Membrane Tolqra Complex In Cymal-6-Neopentyl Glycol Detergent Micelles
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: PROTON TRANSPORT |
 |
Cryo-Em Structure Of The Pma1 With Ordered N-Terminal Extension In The Activated State
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: PROTON TRANSPORT |
 |
Cryo-Em Structure Of The Pma1 With Ordered N-Terminal Extension In The Autoinhibited State
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: PROTON TRANSPORT |
 |
Crystal Structure Of The Light-Driven Proton Pump Heimdallarchaeial Rhodopsin Heimdallr1
Organism: Candidatus heimdallarchaeota archaeon
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: PROTON TRANSPORT Ligands: RET, OLC, NO3 |
 |
Organism: Lama glama, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: PROTON TRANSPORT |
 |
Organism: Lama glama, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: PROTON TRANSPORT |
 |
Organism: Lama glama, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Resolution:3.70 Å Release Date: 2025-02-05 Classification: PROTON TRANSPORT |
 |
Laser Excitation Effects On Br: Reprocessed Dark Dataset Recorded From Nango Et Al.
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Reprocessed Extrapolated 16 Ns Light Dataset Recorded At 0.04 Gw/Cm2 From Nango Et Al.
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Extrapolated 6 Ps Light Dataset Recorded At 342 Gw/Cm2 At Swissfel
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |

