Search Count: 459
 |
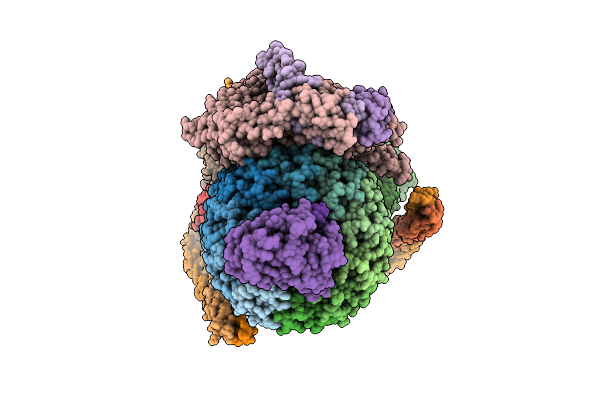
Human V-Atpase Vo Subcomplex (Containing Subunit Isoform A4) Bound To Nanobody And Inhibitor
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN Ligands: NAG, CLR, POV, A1A4Q, WJP |
 |
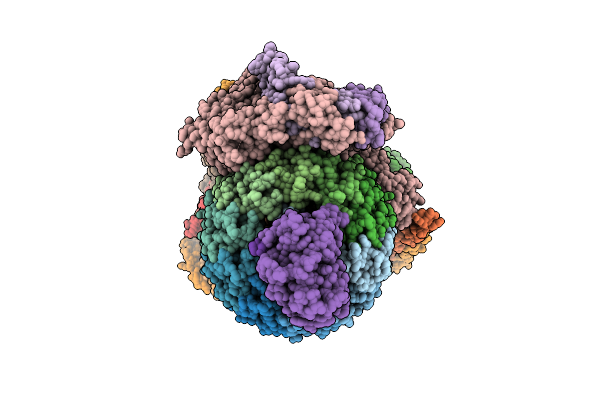
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN Ligands: ADP |
 |
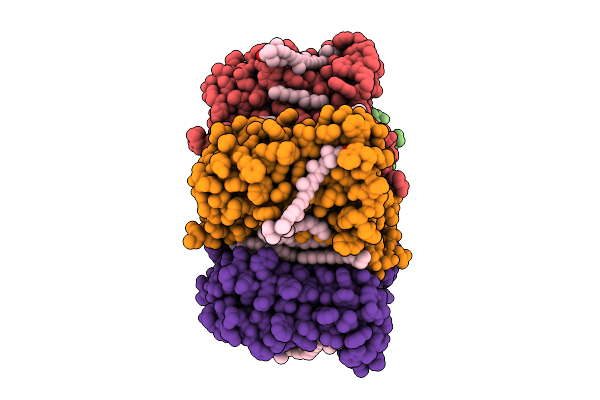
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN Ligands: ADP |
 |
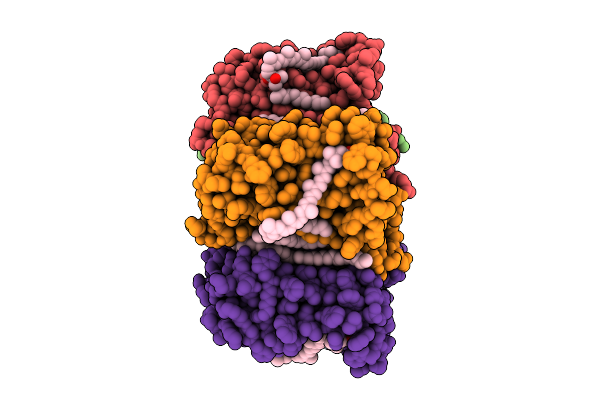
Cryo-Em Structure Of The Zeaxanthin-Bound Light-Driven Proton Pumping Rhodopsin, Nm-R1
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, R16, D12, K3I |
 |
Cryo-Em Structure Of The Myxol-Bound Light-Driven Proton Pumping Rhodopsin, Nm-R1
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, R16, D12, A1L4O |
 |
Cryo-Em Structure Of The Light-Driven Proton Pump Pspr In Detergent Micelle
Organism: Candidatus pseudothioglobus sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA, RET, LMT |
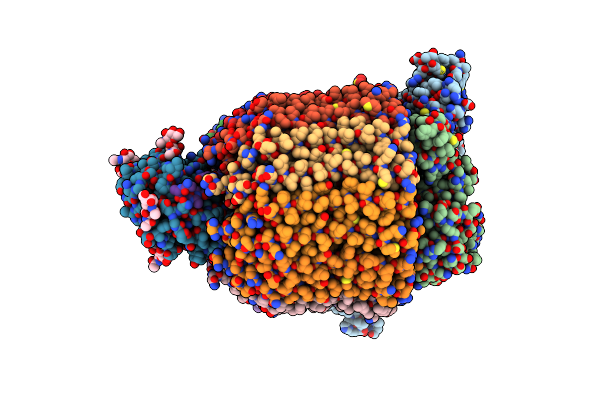 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN |
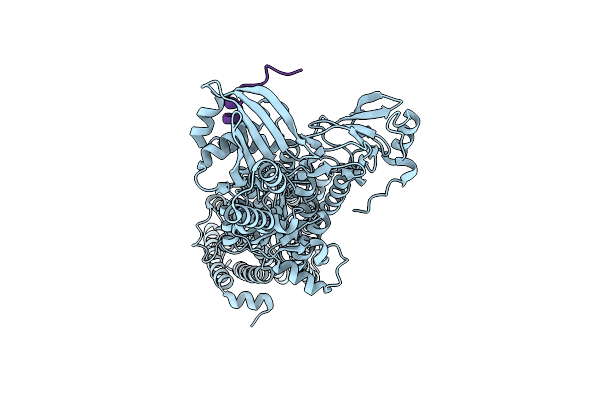 |
Cryo-Em Structure Of The Pma1 With Ordered N-Terminal Extension In The Activated State
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: PROTON TRANSPORT |
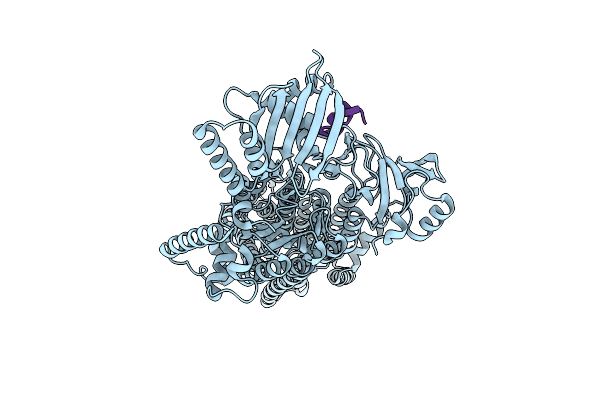 |
Cryo-Em Structure Of The Pma1 With Ordered N-Terminal Extension In The Autoinhibited State
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Resolution:3.52 Å Release Date: 2025-06-04 Classification: PROTON TRANSPORT |
 |
Crystal Structure Of The Light-Driven Proton Pump Heimdallarchaeial Rhodopsin Heimdallr1
Organism: Candidatus heimdallarchaeota archaeon
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-04-02 Classification: PROTON TRANSPORT Ligands: RET, OLC, NO3 |
 |
Organism: Lama glama, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: PROTON TRANSPORT |
 |
Organism: Lama glama, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: PROTON TRANSPORT |
 |
Organism: Lama glama, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Resolution:3.70 Å Release Date: 2025-02-05 Classification: PROTON TRANSPORT |
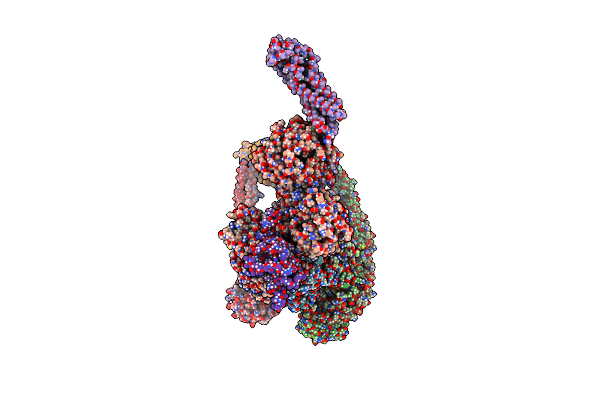 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: HYDROLASE Ligands: MG, ADP |
 |
Organism: Haloquadratum walsbyi dsm 16790
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-10-02 Classification: MEMBRANE PROTEIN Ligands: GOL, 1PE, OLC, RET |
 |
Organism: Uncultured gammaproteobacteria bacterium
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: PROTON TRANSPORT Ligands: DKA |
 |
Organism: Uncultured gammaproteobacteria bacterium
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: PROTON TRANSPORT Ligands: RET |
 |
Organism: Uncultured gammaproteobacteria bacterium
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: PROTON TRANSPORT Ligands: RET |
 |
Organism: Legionella pneumophila subsp. pneumophila str. philadelphia 1, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: PROTON TRANSPORT Ligands: ADP |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: PROTON TRANSPORT Ligands: PC1, WJP, NAG, LP3, PTY, CLR |

