Search Count: 617
All
Selected
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2022-07-06 Classification: SUGAR BINDING PROTEIN Ligands: CL |
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-07-06 Classification: SUGAR BINDING PROTEIN Ligands: FUL, CL |
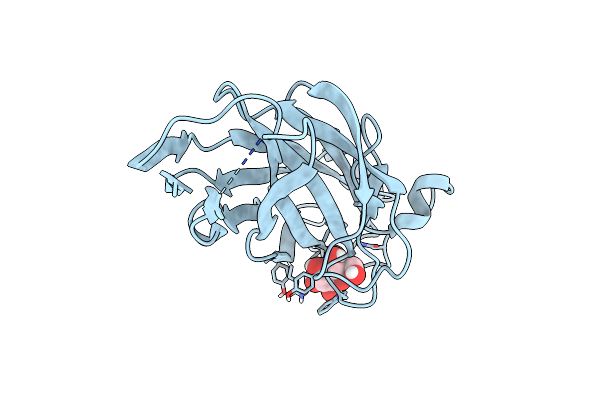 |
Crystal Structure Of The Ucad Lectin-Binding Domain In Complex With Glucose
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2022-07-06 Classification: SUGAR BINDING PROTEIN Ligands: BGC, CL |
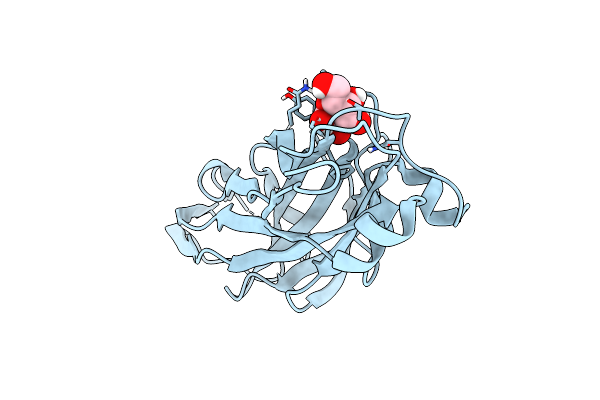 |
Crystal Structure Of The Ucad Lectin-Binding Domain In Complex With Galactose
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2022-07-06 Classification: SUGAR BINDING PROTEIN Ligands: GLA, CL |
 |
X-Ray Structure Of The Zn-Dependent Receptor-Binding Domain Of Proteus Mirabilis Mr/P Fimbrial Adhesin Mrph
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2020-08-19 Classification: CELL ADHESION Ligands: ZN, TLA |
 |
X-Ray Structure Of The Zn-Dependent Receptor-Binding Domain Of Proteus Mirabilis Mr/P Fimbrial Adhesin Mrph
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-08-19 Classification: CELL ADHESION Ligands: ZN, GLU |
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-08-05 Classification: TOXIN |
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2020-08-05 Classification: TOXIN Ligands: GOL |
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2020-08-05 Classification: TOXIN |
 |
Crystal Structure Of The Acyl-Carrier-Protein Udp-N-Acetylglucosamine O-Acyltransferase Lpxa From Proteus Mirabilis
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2020-01-29 Classification: TRANSFERASE Ligands: SO4, PO3 |
 |
Crystal Structure Of Aminoglycoside-Resistance Methyltransferase Rmtc Bound To S-Adenosylhomocysteine (Sah)
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:3.14 Å Release Date: 2019-10-16 Classification: TRANSFERASE Ligands: SAH |
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2019-07-24 Classification: HYDROLASE Ligands: CA, MPD |
 |
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2019-06-26 Classification: LYASE Ligands: SO4 |
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2019-03-06 Classification: ISOMERASE |
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-03-06 Classification: OXIDOREDUCTASE |
 |
Organism: Proteus mirabilis hi4320
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2018-11-07 Classification: CELL ADHESION Ligands: GOL, PO4 |
 |
Receptor-Binding Domain Of Proteus Mirabilis Uroepithelial Cell Adhesin Ucad21-211
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-11-07 Classification: CELL ADHESION Ligands: CO |
 |
Receptor-Binding Domain Of Proteus Mirabilis Uroepithelial Cell Adhesin Ucad21-217
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-11-07 Classification: CELL ADHESION Ligands: SO4 |
 |
Substrate-Bound Outward-Open State Of A Na+-Coupled Sialic Acid Symporter Reveals A Novel Na+-Site
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-04-04 Classification: MEMBRANE PROTEIN |
 |
Substrate-Bound Outward-Open State Of A Na+-Coupled Sialic Acid Symporter Reveals A Novel Na+-Site
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2018-04-04 Classification: MEMBRANE PROTEIN |

