Search Count: 8,022
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: ISOMERASE Ligands: 9CK, EDO, 1PE, PGE, PO4, NA |
 |
Crystal Structure Of Streptomyces Sp. Sulfotransferase Cpz8 Involved In Caprazamycins Synthesis
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: PO4, MG |
 |
Discovery Of A Bifunctional Pkmyt1-Targeting Protac Empowered By Ai-Generation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: PROTEIN BINDING Ligands: A1EMX, PO4, CL |
 |
Organism: Bacillus sp. ps3
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: ELECTRON TRANSPORT Ligands: MG, ATP, PO4, ADP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: IMMUNE SYSTEM Ligands: PO4 |
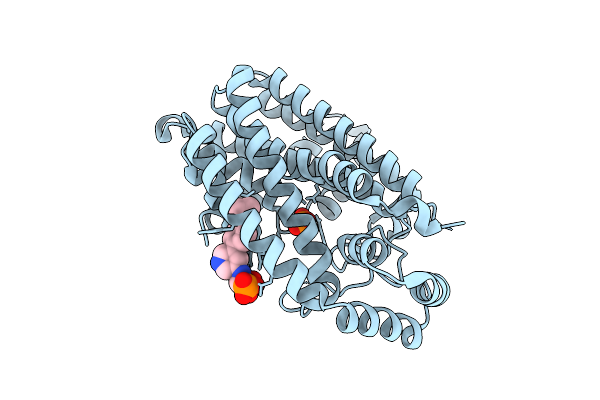 |
Crystal Structure Of Human Farnesyl Pyrophosphate Synthase In Complex With A Cryptic Pocket Ligand, Jds05120
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: PO4, JD5 |
 |
Crystal Structure Of Bip Atpase Domain In Complex With Cdnf C-Terminal Domain At 1.65 Angstroms Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: CHAPERONE Ligands: MG, CL, PO4, GOL |
 |
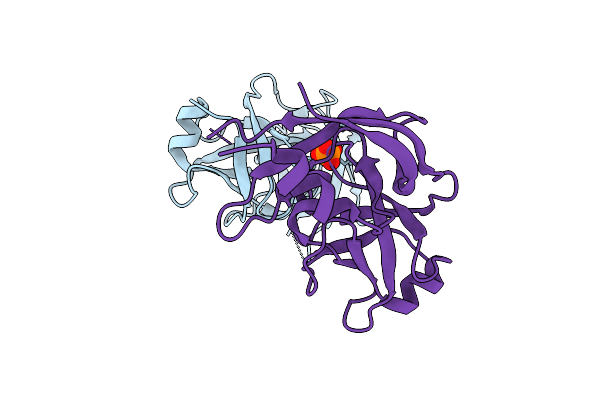
Cereblon Isoform 4 From Magnetospirillum Gryphiswaldense In Complex With Glutarimide Based Compound 2J
Organism: Magnetospirillum gryphiswaldense
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: SIGNALING PROTEIN Ligands: ZN, A1IW9, PO4 |
 |
Cereblon Isoform 4 From Magnetospirillum Gryphiswaldense In Complex With Glutarimide Based Compound 2S
Organism: Magnetospirillum gryphiswaldense
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: SIGNALING PROTEIN Ligands: ZN, A1IW7, PO4, A1I4Q |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: PLANT PROTEIN Ligands: PO4 |
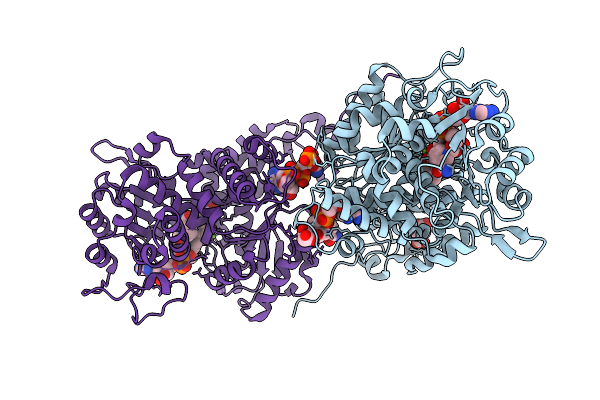 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: OXIDOREDUCTASE Ligands: MG, A1AE5, SIN, NAD, PO4 |
 |
Organism: Rhizobium tropici ciat 899
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: A1CCU, GLY, GOL, PO4 |
 |
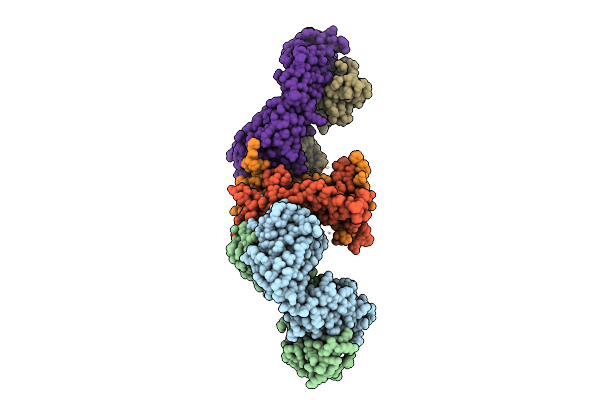
Structure Of The 70S-Ef-G(P610L)-Gdp-Pi Ribosome Complex With Trnas In Hybrid State 2 (H2-Ef-G(P610L)-Gdp-Pi)
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME Ligands: MG, ZN, NA, AM2, GDP, PO4 |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: CYTOKINE Ligands: ZN, PO4 |
 |
Cryo-Em Structure Of The V1 Domain Of V/A-Atpase In Liposomes Under No Pmf Condition, State1W
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MOTOR PROTEIN Ligands: MG, ADP, PO4 |
 |
Cryo-Em Structure Of The V1 Domain Of V/A-Atpase In Liposomes Under No Pmf Condition, State1M
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MOTOR PROTEIN Ligands: MG, ADP, PO4 |
 |
Cryo-Em Structure Of The V1 Domain Of V/A-Atpase In Liposomes Under No Pmf Condition, State1N
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MOTOR PROTEIN Ligands: MG, ADP, PO4 |
 |
Cryo-Em Structure Of The V1 Domain Of V/A-Atpase In Liposomes Under Pmf Condition, State1W
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MOTOR PROTEIN Ligands: MG, ATP, ADP, PO4 |
 |
Cryo-Em Structure Of The V1 Domain Of V/A-Atpase In Liposomes Under Pmf Condition, State1M
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MOTOR PROTEIN Ligands: MG, ATP, ADP, PO4 |
 |
Cryo-Em Structure Of The V1 Domain Of V/A-Atpase In Liposomes Under Pmf Condition, State1N
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MOTOR PROTEIN Ligands: MG, ATP, ADP, PO4 |

