Search Count: 31
 |
Crystal Structure Of Penicillin Binding Protein 5 (Pbp5) Papapap Variant Penicillin Bound Form From Enterococcus Faecium
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-11-15 Classification: ANTIBIOTIC Ligands: PNM, SO4 |
 |
Crystal Structure Of Penicillin Binding Protein 5 (Pbp5) Poly-Gly Variant Penicillin Bound Form From Enterococcus Faecium
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2023-11-15 Classification: ANTIBIOTIC Ligands: PNM, SO4 |
 |
Crystal Structure Of Penicillin Binding Protein 5 (Pbp5) T485M Variant In The Penicillin Bound Form From Enterococcus Faecium
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:3.59 Å Release Date: 2023-07-05 Classification: ANTIBIOTIC Ligands: PNM, SO4 |
 |
Crystal Structure Of Penicillin Binding Protein 5 (Pbp5) S466 Insertion Variant Penicillin Bound Form From Enterococcus Faecium
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-07-05 Classification: ANTIBIOTIC Ligands: SO4, PNM |
 |
Crystal Structure Of Penicillin Binding Protein 5 (Pbp5) T485A Variant Penicillin Bound Form From Enterococcus Faecium
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2023-07-05 Classification: ANTIBIOTIC Ligands: PNM, SO4 |
 |
Crystal Structure Of Penicillin Binding Protein 5 (Pbp5) T485A Variant With S466 Insertion Penicillin Bound Form From Enterococcus Faecium
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2023-07-05 Classification: ANTIBIOTIC Ligands: PNM, SO4 |
 |
Crystal Structure Of Penicillin Binding Protein 5 (Pbp5) R464A Variant Penicillin Bound Form From Enterococcus Faecium
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2023-07-05 Classification: ANTIBIOTIC Ligands: PNM, SO4 |
 |
Crystal Structure Of Penicillin Binding Protein 5 (Pbp5) T485M T499I Variant Penicillin Bound Form From Enterococcus Faecium
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2023-07-05 Classification: ANTIBIOTIC Ligands: PNM, SO4 |
 |
Crystal Structure Of Penicillin Binding Protein 5 (Pbp5) T485M T499I V629E Variant Penicillin Bound Form From Enterococcus Faecium
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-07-05 Classification: ANTIBIOTIC Ligands: PNM, SO4 |
 |
Crystal Structure Of Penicillin-Binding Protein 1 (Pbp1) From Staphylococcus Aureus In Complex With Penicillin G
Organism: Staphylococcus aureus subsp. aureus col
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: CIT, SO4, PNM |
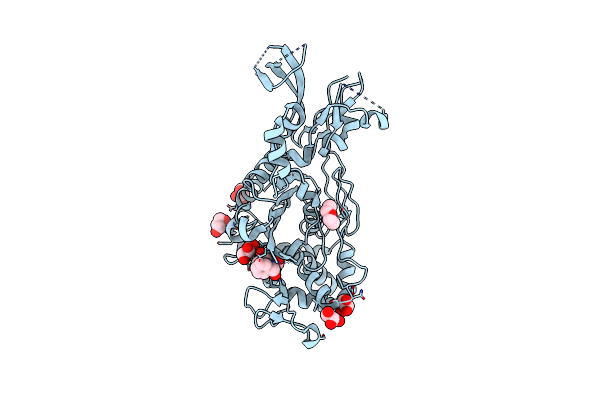 |
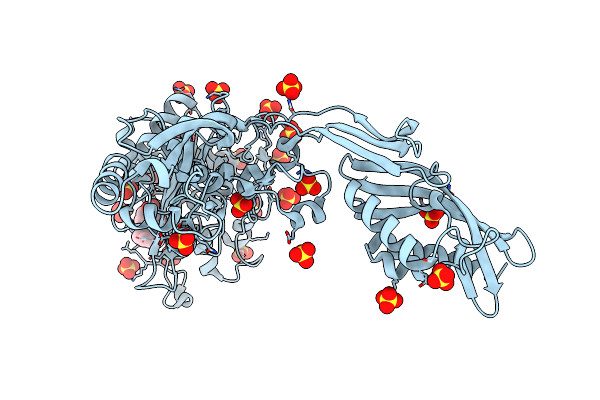
Crystal Structure Of Penicillin-Binding Protein 4 (Pbp4) From Enterococcus Faecalis In The Benzylpenicillin Bound Form.
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2018-10-31 Classification: HYDROLASE Ligands: GOL, CL, PEG, PNM |
 |
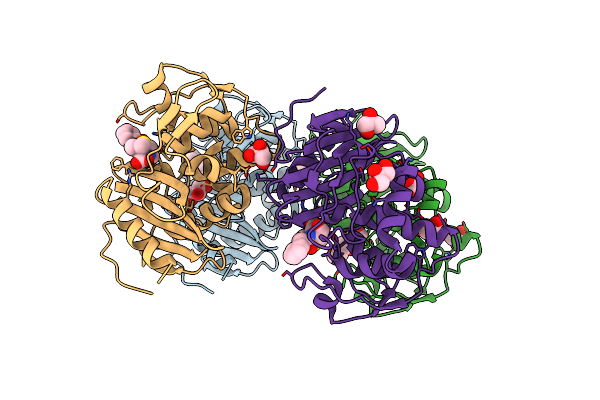
Crystal Structure Of Penicillin Binding Protein 5 (Pbp5) From Enterococcus Faecium In The Benzylpenicilin-Bound Form
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2018-10-31 Classification: protein binding/antibiotic Ligands: PNM, SO4 |
 |
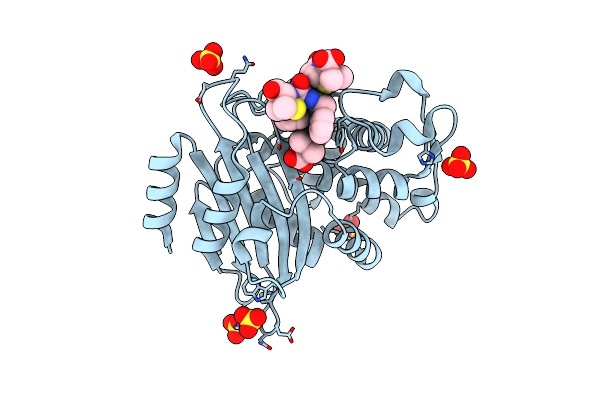
Crystal Structure Of Penicillin-Binding Protein D2 From Listeria Monocytogenes In The Penicillin G Bound Form
Organism: Listeria monocytogenes egd-e
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-07-25 Classification: ANTIBIOTIC Ligands: PNM, GOL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2017-03-01 Classification: HYDROLASE Ligands: SO4, PNM, PNN |
 |
Penicillin G Acylated Bd3459 Predatory Endopeptidase From Bdellovibrio Bacteriovorus In Complex With Immunity Protein Bd3460
Organism: Bdellovibrio bacteriovorus hd100
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2015-12-09 Classification: HYDROLASE Ligands: PNM |
 |
Crystal Structure Of New Delhi Metallo-Beta-Lactamase-1 Mutant M67V Complexed With Hydrolyzed Penicillin G
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-09-24 Classification: HYDROLASE/ANTIBIOTIC Ligands: ZN, PNM, CL |
 |
Structure Of A Putative Peptidoglycan Glycosyltransferase From Atopobium Parvulum In Complex With Penicillin G
Organism: Atopobium parvulum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-10-30 Classification: Transferase/Antibiotic Ligands: PNM |
 |
Structure Of Penicillin-Binding Protein A From M. Tuberculosis: Penicillin G Acyl-Enzyme Complex
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-10-24 Classification: Penicillin-binding protein/Antibiotic Ligands: PNM |
 |
Crystal Structure Of Acinetobacter Baumannii Pbp1A In Complex With Penicillin G
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-12-14 Classification: Penicillin-binding protein/ANTIBIOTIC Ligands: PNM |
 |
Crystal Structure Of Fluorophore-Labeled Class A -Lactamase Penp-E166Cb In Complex With Penicillin G
Organism: Bacillus licheniformis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-03-02 Classification: HYDROLASE Ligands: PNM |

