Search Count: 40
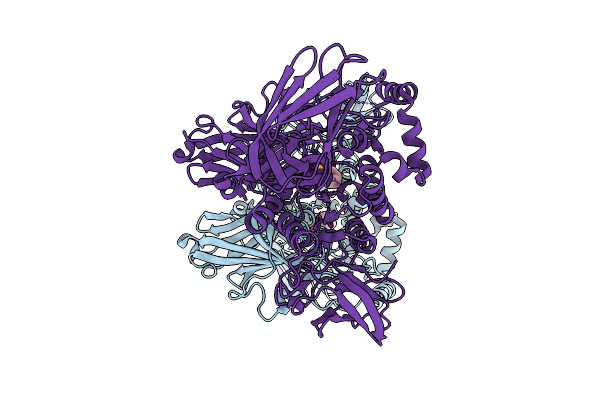 |
Crystal Structure Of A Ce20 Carbohydrate Acetylesterase From Pedobacter Psychrotolerans (Ppce20_Ii), With Ancillary Domain
Organism: Pedobacter psychrotolerans
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-08-13 Classification: HYDROLASE Ligands: PGE, MG, PMS |
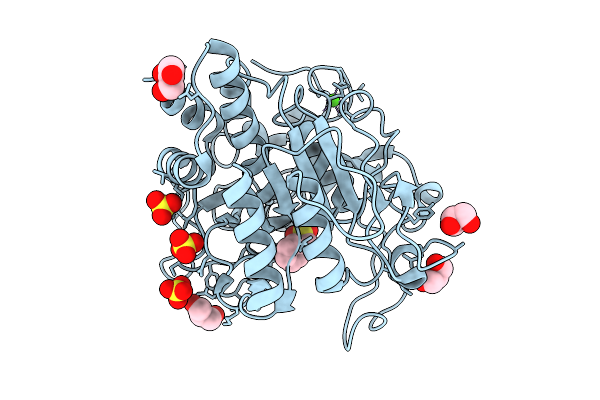 |
Stmpr1, Stenotrophomonas Maltophilia Protease 1, 36 Kda Alkine Serine Protease In Complex With Pmsf
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-08-06 Classification: HYDROLASE Ligands: GOL, PMS, SO4, CA |
 |
Organism: Sipunculus nudus
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2025-04-02 Classification: BLOOD CLOTTING Ligands: SO4, PMS |
 |
Organism: Sipunculus nudus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-04-02 Classification: BLOOD CLOTTING Ligands: PMS, SO4, GOL, 1PE |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2025-02-05 Classification: HYDROLASE Ligands: SO4, PMS |
 |
Crystal Structure Of The Thermotoga Maritima Cold Shock Protein Mutant Est#13 In Complex With Aacest
Organism: Alicyclobacillus acidocaldarius, Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-15 Classification: PROTEIN BINDING Ligands: PMS, PEG, GOL |
 |
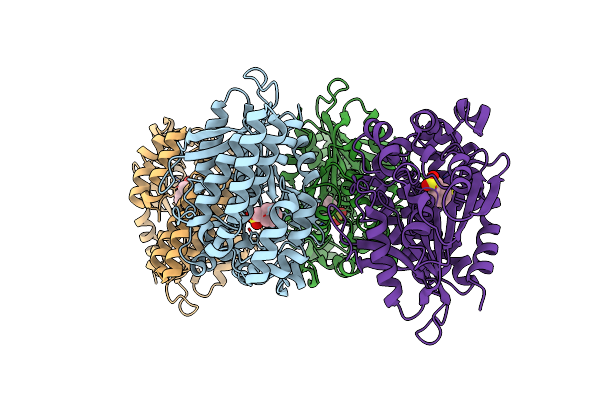
Crystal Structure Of A Novel Pu Plastic Degradation Urethanase Umg-Sp2 From Uncultured Bacterium
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-09-18 Classification: HYDROLASE Ligands: GOL, SO4, PMS |
 |
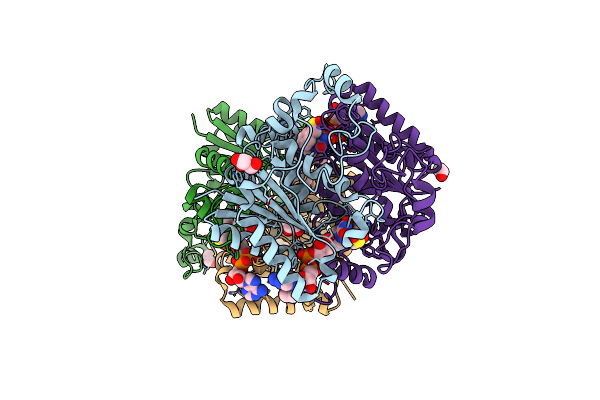
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-03-13 Classification: LYASE Ligands: PMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-11-01 Classification: FLAVOPROTEIN Ligands: FAD, PMS, GOL, ACE |
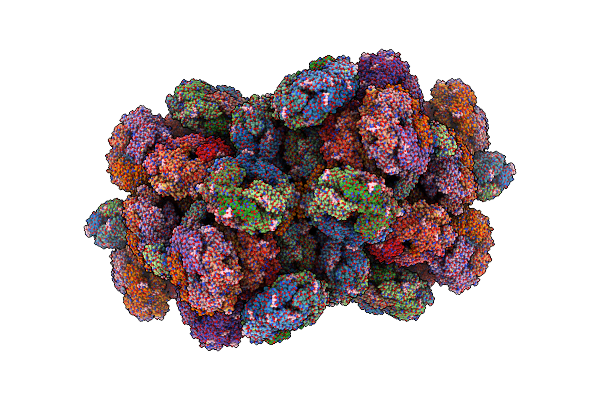 |
Structure Of The Phycobilisome From The Red Alga Porphyridium Purpureum In Middle Light
Organism: Porphyridium purpureum
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: PHOTOSYNTHESIS Ligands: PEB, PUB, CYC, PMS |
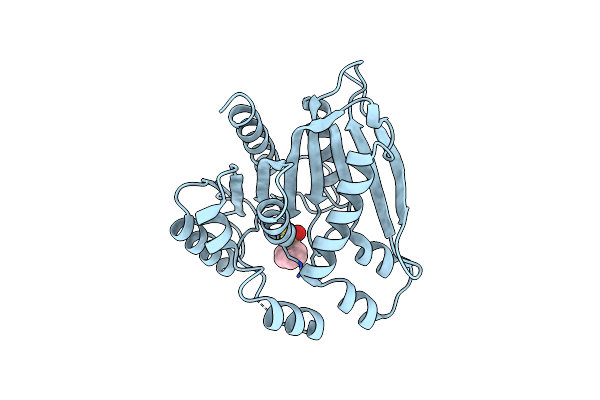 |
Crystal Structure Of Psest3 Complexed With Phenylmethylsulfonyl Fluoride (Pmsf)
Organism: Paenibacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2022-08-31 Classification: HYDROLASE Ligands: PMS |
 |
Crystal Structure Of An Engineered Variant Of Single-Chain Penicillin G Acylase From Kluyvera Cryocrescens (A1-Ac Rd3Chis)
Organism: Kluyvera cryocrescens
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2021-11-17 Classification: HYDROLASE Ligands: CA, PMS |
 |
Crystal Structure Of Mycobacterium Tuberculosis Htra1 (Rv1223) In Regulated Conformation
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2018-12-19 Classification: MEMBRANE PROTEIN Ligands: PMS |
 |
Crystal Structure Of Lysophospholipase A2 Conjugated With Phenylmethylsulfonyl Fluoride
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-11-14 Classification: PROTEIN BINDING Ligands: PMS |
 |
Crystal Structure Of A Novel Pyrethroid Hydrolase From Sphingobium Faniae Jz-2
Organism: Sphingobium faniae
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2018-08-08 Classification: HYDROLASE Ligands: PMS, SO4 |
 |
Organism: Penicillium cyclopium
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-02-28 Classification: HYDROLASE Ligands: CA, PMS |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-07-26 Classification: HYDROLASE Ligands: PMS |
 |
Crystal Structure Of Low Molecular Weight Protein Tyrosine Phosphatase Isoform A Complexed With Phenylmethanesulfonic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-07-15 Classification: HYDROLASE Ligands: GOL, SO4, PMS |
 |
Crystal Structure Of An Esterase From The Bacterial Hormone-Sensitive Lipase (Hsl) Family
Organism: Environmental samples
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-03-25 Classification: HYDROLASE Ligands: PMS |
 |
Structural And Functional Studies On A Thermostable Polyethylene Terephthalate Degrading Hydrolase From Thermobifida Fusca
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2014-06-25 Classification: HYDROLASE Ligands: PMS, SO4 |

