Search Count: 126
 |
C387S Variant Of D-Ornithine/D-Lysine Decarboxylase Complexed With Pmp And 4-Guanidinobutanal
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: LYASE Ligands: DMS, NA, PMP, A1AZK, ACT, CL |
 |
Crystal Structure Of A Heterooligomeric Aminotransferase From Serratia Sp. Atcc 39006, Pmp-Bound Form
Organism: Serratia sp. atcc 39006
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: PMP |
 |
Organism: Micromonospora echinospora
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2024-09-11 Classification: ANTIBIOTIC Ligands: PMP, GET, CL |
 |
Organism: Micromonospora echinospora
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-09-11 Classification: ANTIBIOTIC Ligands: GOL, PMP, CL |
 |
Organism: Micromonospora echinospora
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2024-09-11 Classification: BIOSYNTHETIC PROTEIN Ligands: 827, PMP, CL |
 |
Crystal Structure Of D-Amino Acid Aminotransferase From Blastococcus Saxobsidens In Pmp Form
Organism: Blastococcus saxobsidens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-07-10 Classification: TRANSFERASE Ligands: PMP, EDO, NA |
 |
Crystal Structure Of D-Amino Acid Aminotransferase From Aminobacterium Colombiense Point Mutant E113A Complexed With D-Glutamate
Organism: Aminobacterium colombiense
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-04-10 Classification: TRANSFERASE Ligands: EDO, NO3, PMP, PW0 |
 |
Crystal Structure Of The Pcryo_0618 Aminotransferase From Psychrobacter Cryohalolentis K5 In The Presence Of Pmp And Glutamate
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-01-31 Classification: TRANSFERASE Ligands: PMP, A1ADI, CL, EDO, MG, PDG, NA |
 |
Structure Of S. Hygroscopicus Aminotransferase Mppq Complexed With Pyridoxamine 5'-Phosphate (Pmp)
Organism: Streptomyces hygroscopicus
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2023-11-22 Classification: BIOSYNTHETIC PROTEIN Ligands: PMP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2023-03-29 Classification: TRANSFERASE Ligands: SO4, PMP |
 |
Crystal Structure Of The Human Phosphoserine Aminotransferase (Psat) In Complex With O-Phosphoserine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2023-03-29 Classification: TRANSFERASE Ligands: E1U, SEP, PMP, SO4 |
 |
1.55 A X-Ray Crystallographic Structure Of Saph From Streptomyces Sp. (Hph0547) Involved In Pseudouridimycin Biosynthesis
Organism: Streptomyces sp. hph0547
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-02-15 Classification: BIOSYNTHETIC PROTEIN Ligands: PMP, GOL, K |
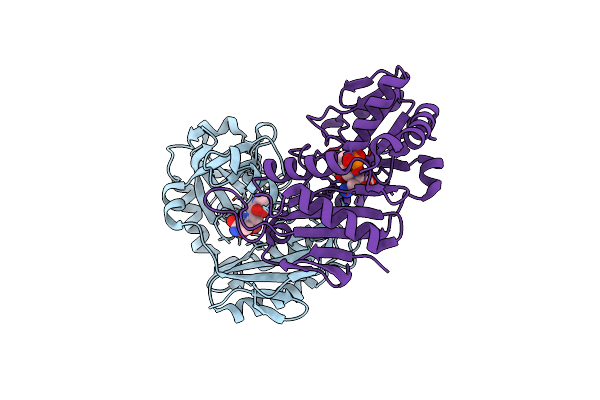 |
Crystal Structure Of D-Amino Acid Aminotransferase From Aminobacterium Colombiense Complexed With D-Cycloserine
Organism: Aminobacterium colombiense
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-11-16 Classification: TRANSFERASE Ligands: LCS, PMP, M7L |
 |
Crystal Structure Of D-Amino Acid Aminotrensferase From Aminobacterium Colombiense Complexed With D-Glutamate
Organism: Aminobacterium colombiense
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-11-16 Classification: TRANSFERASE Ligands: PW0, PMP |
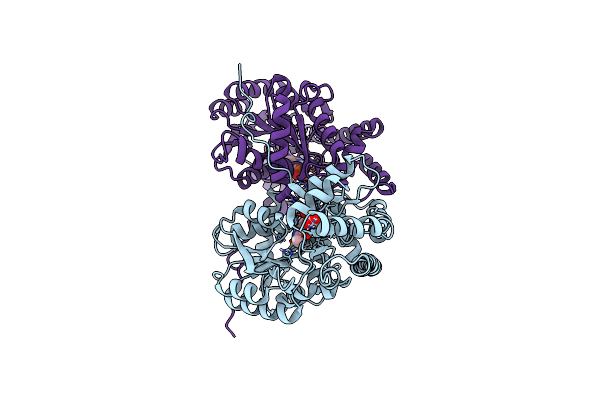 |
Joint X-Ray/Neutron Structure Of Aspastate Aminotransferase (Aat) In Complex With Pyridoxamine 5'-Phosphate (Pmp)
Organism: Sus scrofa
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.70 Å, 2.22 Å Release Date: 2022-09-28 Classification: TRANSFERASE Ligands: PLA, PMP, DOD |
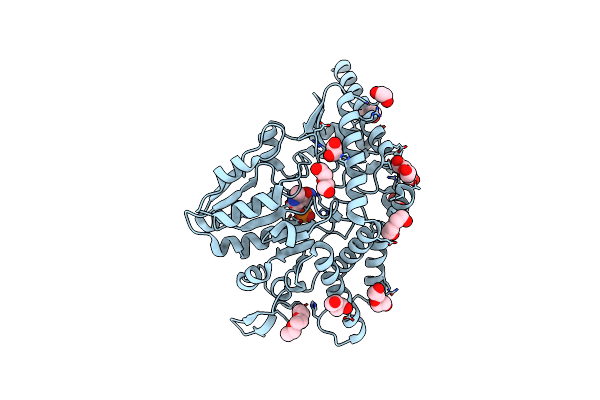 |
Crystal Structure Of Pmp-Bound Form Of Cysteine Desulfurase Sufs C361A From Bacillus Subtilis
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-03-02 Classification: BIOSYNTHETIC PROTEIN Ligands: PMP, EDO, PEG, PG4 |
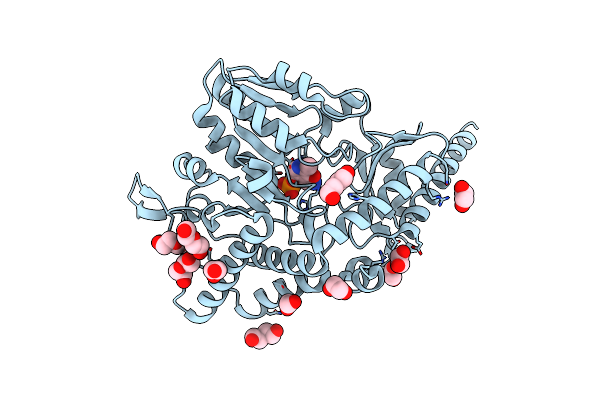 |
Crystal Structure Of Pmp-Bound Form Of Cysteine Desulfurase Sufs R376A From Bacillus Subtilis In D-Cycloserine-Inhibition
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2022-03-02 Classification: BIOSYNTHETIC PROTEIN Ligands: PMP, PG4, PEG, EDO |
 |
Crystal Structure Of Pmp-Bound Form Of Cysteine Desulfurase Sufs R376A From Bacillus Subtilis In L-Cycloserine-Inhibition
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2022-03-02 Classification: BIOSYNTHETIC PROTEIN Ligands: PEG, PMP, EDO |
 |
Thermostable Omega Transaminase Pjta-R6 Variant W58M/F86L/R417L Engineered For Asymmetric Synthesis Of Enantiopure Bulky Amines
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-09-01 Classification: TRANSFERASE Ligands: PMP, SIN |
 |
Crystal Structure Of Pmp-Bound Form Of Cysteine Desulfurase Sufs From Bacillus Subtilis
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2021-06-30 Classification: BIOSYNTHETIC PROTEIN Ligands: PEG, PMP |

