Search Count: 1,348
 |
Crystal Structure Of A Transaminase Pata From Pseudonocardia Ammonioxydans In Complex With Plp And Llp
Organism: Pseudonocardia ammonioxydans
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: PLP |
 |
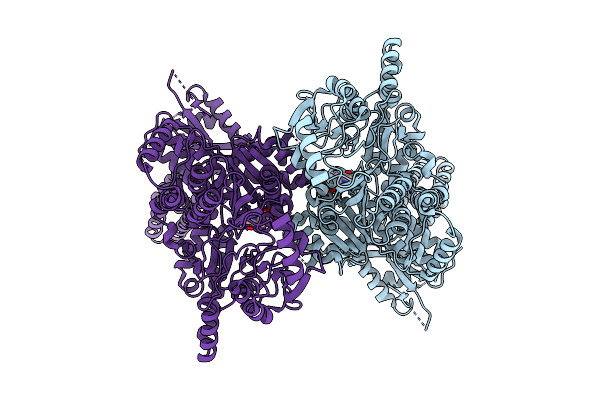
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: PLP |
 |
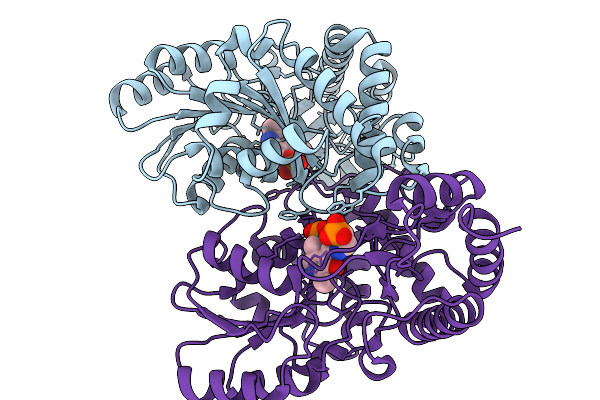
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: LYASE Ligands: PLP |
 |
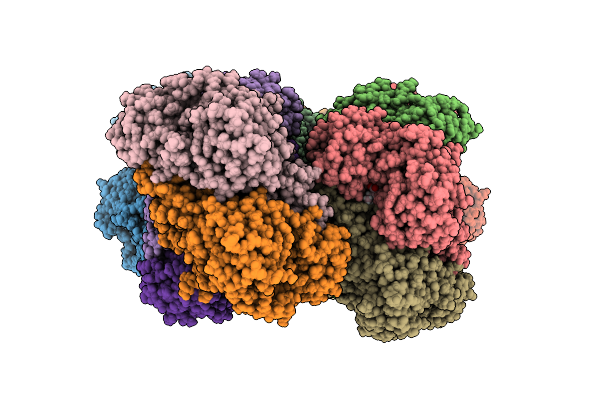
Structure Of The K193M Mutant Of Transaminase Phnw From Vibrio Vulnificus In Complex With Plp And Aep
Organism: Vibrio vulnificus nbrc 15645 = atcc 27562
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSFERASE Ligands: PLP, P7I, CL |
 |
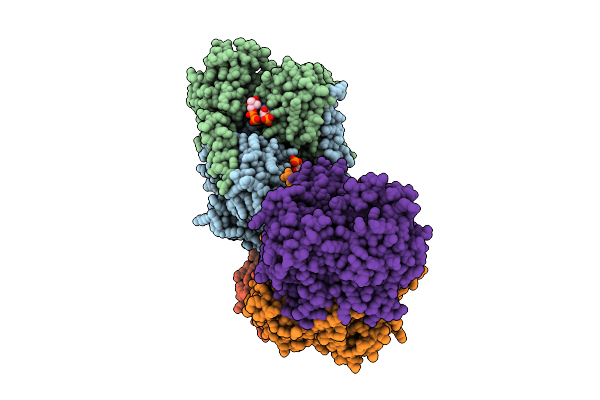
Crystal Structure Of Methionine Gamma-Lyase From Brevibacterium Sandarakinum In Complex With Plp And Norleucine At Ph 8.5
Organism: Brevibacterium sandarakinum
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: LYASE Ligands: NLE, PY6, PEG, PLP |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: PLP, SCA, MG |
 |
Crystal Structure Of Capa2 From Alistipes Finegoldii In Complex With Plp Cofactor
Organism: Alistipes finegoldii dsm 17242
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: BIOSYNTHETIC PROTEIN Ligands: CIT, PLP |
 |
Organism: Streptomyces sp. bv333
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: TRANSFERASE Ligands: EDO, PLP, SO4 |
 |
The Structure Of Ornithine Decarboxylase From Leishmania Infantum In Complex With Plp And Dfmo
Organism: Leishmania infantum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: LYASE Ligands: PLP, DMO |
 |
Structure Of Aminotransferase From Mycolicibacterium Neoaurum In Complex With Plp
Organism: Mycolicibacterium neoaurum
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: TRANSFERASE Ligands: PLP |
 |
Organism: Parageobacillus caldoxylosilyticus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: PLP |
 |
X-Ray Structure Of Human Holo Aromatic L-Amino Acid Decarboxylase (Aadc) Complex With Carbidopa At Physiological Ph
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN Ligands: PLP, 142, PG4 |
 |
Organism: Acidithiobacillus
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2025-05-14 Classification: LYASE Ligands: PLP |
 |
Structure Of Human Glutamate Oxaloacetate Transaminase 1 (Got1) Mutant - P15R
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2025-04-16 Classification: TRANSFERASE Ligands: PLP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: PLP |
 |
Crystal Structure Of Ornithine Decarboxylase In Complex With A Novel Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2025-03-12 Classification: CARBOHYDRATE Ligands: A1AQO, PLP |
 |
Crystal Structure Of Ornithine Decarboxylase In Complex With A Novel Inhibitor (10-S)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-12 Classification: CARBOHYDRATE Ligands: PLP, A1AQP |
 |
Crystal Structure Of The Dimeric Transaminase Doed From C. Salexigens Dsm 3043.
Organism: Chromohalobacter israelensis dsm 3043
Method: X-RAY DIFFRACTION Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: GOL, PLP |
 |
Crystal Structure Of Tyrosine Decarboxylase In Complex With The Cofactor Plp And Inhibitor Carbidopa
Organism: Papaver somniferum
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2025-02-12 Classification: LYASE Ligands: PLP, 142 |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: PLP, CL, ALA |

