Search Count: 723
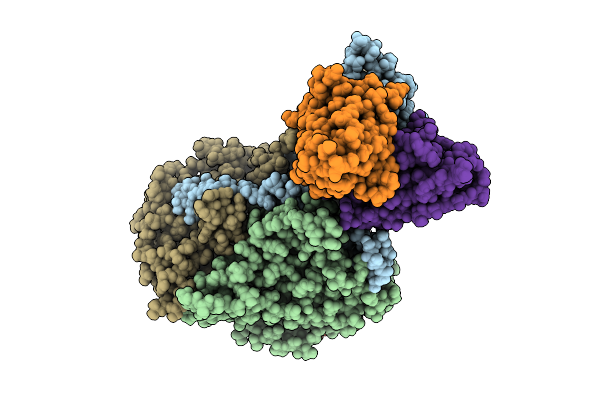 |
Organism: Poliovirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRUS Ligands: PLM |
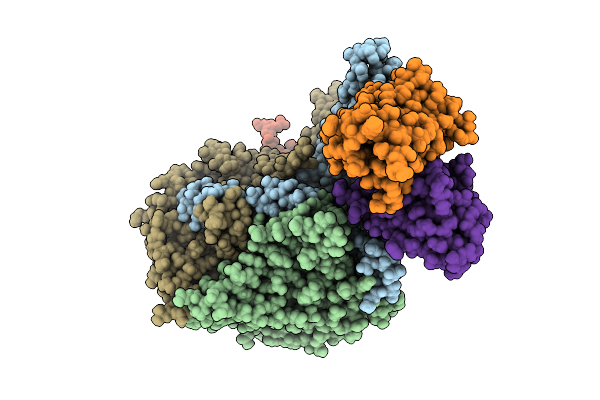 |
Organism: Poliovirus 3, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRUS Ligands: PLM |
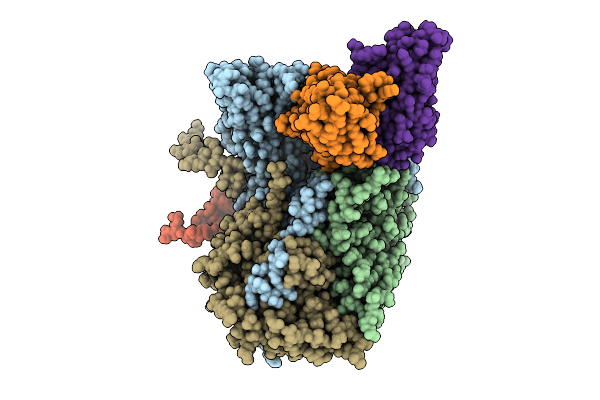 |
Organism: Poliovirus 3, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRUS Ligands: PLM |
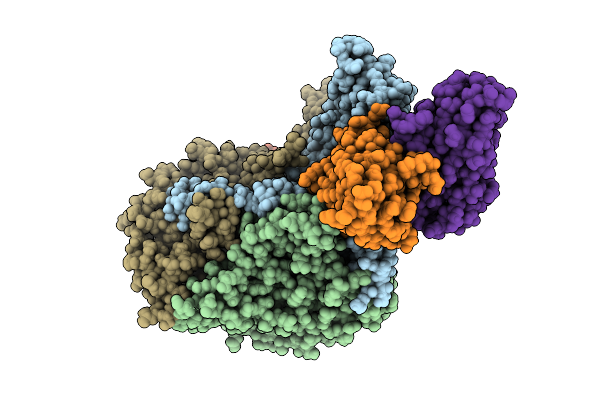 |
Organism: Human poliovirus 1 strain sabin, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRUS Ligands: PLM |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: MG, DD9, D10, HP6, XKP, PEF, PLM, DCR |
 |
Cryo-Em Structure Of Lptdem Complex Containing Shigella Flexneri Lpte And Endogenous E. Coli Lptd And Lptm
Organism: Shigella flexneri, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: PLM, PXS |
 |
Cryo-Em Structure Of Shigella Flexneri Lptde In Complex With Rtp45 Superinfection Exclusion Protein From Rtp Bacteriophage And Endogenous Lptm
Organism: Shigella flexneri, Escherichia phage rtp, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: PLM, PXS |
 |
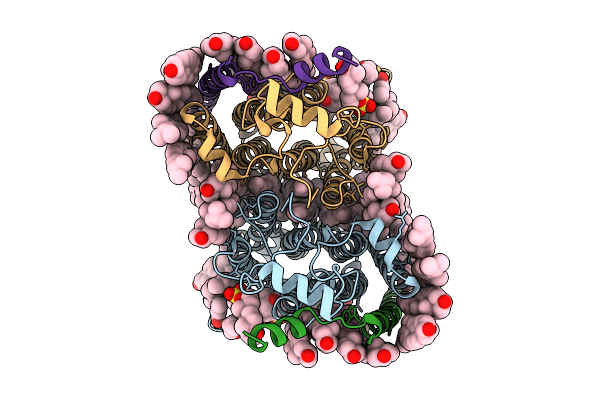
Cryo-Em Structure Of Human Organic Solute Transporter Ost-Alpha/Beta Bound With Tlca
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: CLR, P0E, LPE, NAG, PLM, A1EPX, 76F |
 |
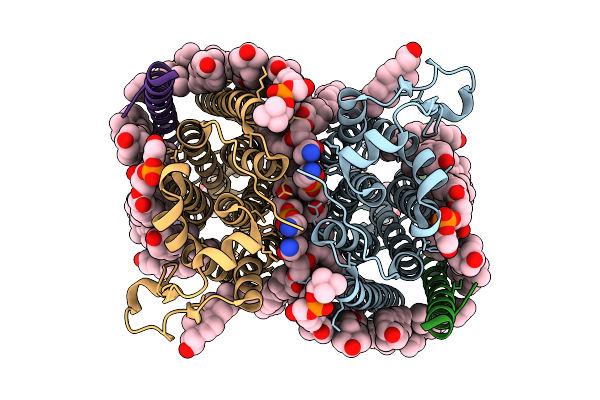
Cryo-Em Structure Of Human Organic Solute Transporter Ost-Alpha/Beta Bound With Dheas
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: 76F, P0E, LPE, CLR, PLM, ZWY |
 |
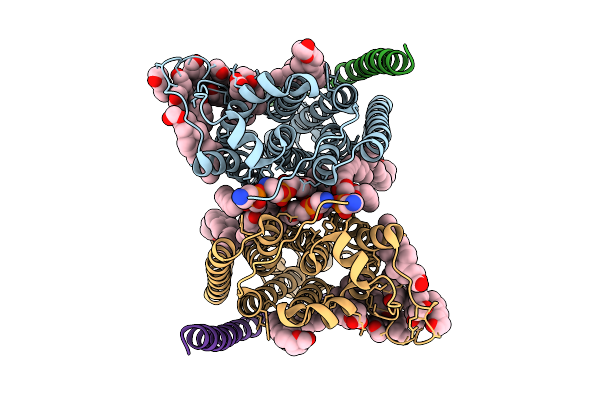
Cryo-Em Structure Of Human Organic Solute Transporter Ost-Alpha/Beta In Apo State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: 76F, P0E, LPE, CLR, NAG, PLM |
 |
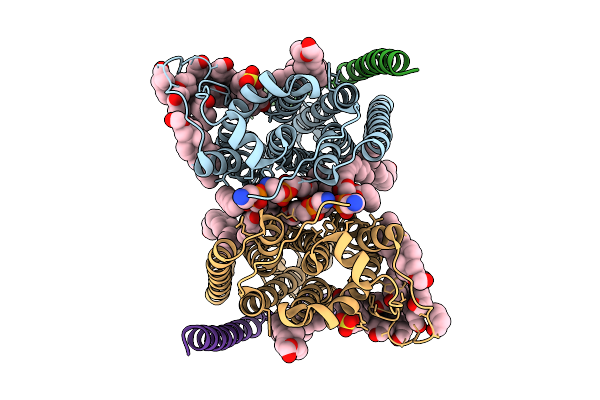
Cryo-Em Structure Of Human Organic Solute Transporter Ostalpha/Beta In Apo State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: TRANSPORT PROTEIN Ligands: PLM, LBN, Y01 |
 |
Cryo-Em Structure Of Human Organic Solute Transporter Ostalpha/Beta In Complex With Dheas
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: TRANSPORT PROTEIN Ligands: PLM, LBN, Y01, ZWY |
 |
Organism: Porphyromonas gingivalis atcc 33277
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: PLM, Z41 |
 |
Organism: Bat coronavirus hku5
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRAL PROTEIN Ligands: PLM, NAG, OLA |
 |
Organism: Bat coronavirus hku5
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRAL PROTEIN Ligands: PLM, NAG |
 |
Cryo-Em Structure Of Shigella Flexneri Lptde Bound By A Bicyclic Peptide Molecule (Compound 16)
Organism: Shigella flexneri, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: LMT, PLM, Z41, A1I4O |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: MEMBRANE PROTEIN Ligands: GDP, ZN, NAG, Y01, PLM, NA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: MEMBRANE PROTEIN Ligands: NAG, GDP, ZN, PLM, ATP |
 |
Cryo-Em Structure Of The Human P2X7 Receptor In The Ub-Alt-P30-Bound Inhibited State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: MEMBRANE PROTEIN Ligands: A1BD8, GDP, ZN, NAG, Y01, PLM |
 |
Cryo-Em Structure Of The Human P2X7 Receptor In The Ub-Mbx-46-Bound Inhibited State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: MEMBRANE PROTEIN Ligands: A1BD9, GDP, ZN, NAG, Y01, PLM, NA |

