Search Count: 754
All
Selected
 |
Organism: Mus musculus, Gallus gallus
Method: ELECTRON MICROSCOPY Release Date: 2026-02-18 Classification: PROTEIN BINDING |
 |
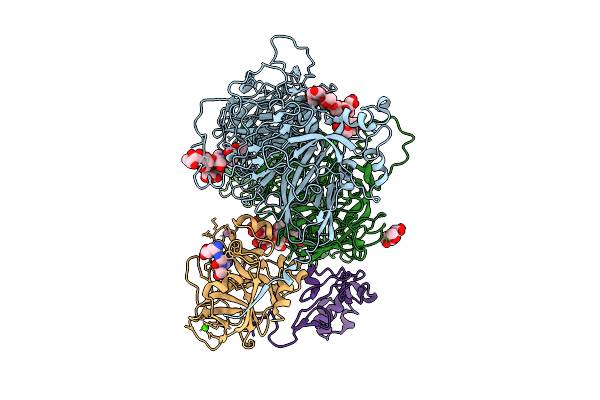
Target Dna-Bound Type I-F3 Tniq-Cascade Of Vibrio Parahaemolyticus In Partial R-Loop State
Organism: Vibrio parahaemolyticus rimd 2210633, Vibrio alginolyticus
Method: ELECTRON MICROSCOPY Resolution:2.68 Å Release Date: 2026-02-11 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: DNA BINDING PROTEIN |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Resolution:2.93 Å Release Date: 2025-12-17 Classification: DNA BINDING PROTEIN/DNA |
 |
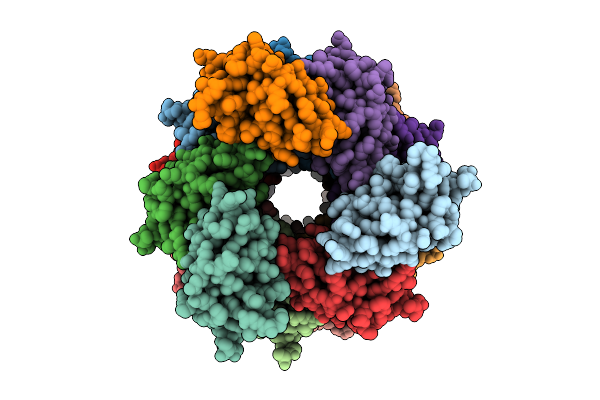
A 3.3 Angstrom Cryo-Em Structure Of An Engineered High-Affinity Human Prothrombinase Complex
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: BLOOD CLOTTING Ligands: CA, NA, 0GJ, CU |
 |
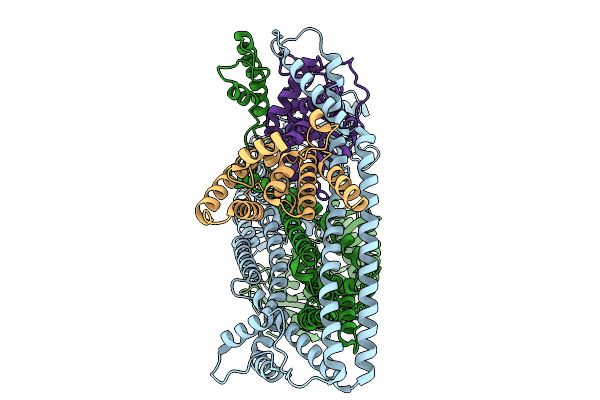
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: IMMUNE SYSTEM |
 |
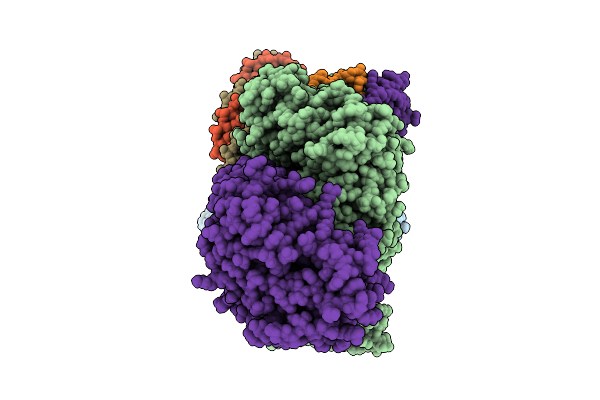
Organism: Salmonella enterica subsp. enterica serovar tennessee
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: IMMUNE SYSTEM |
 |
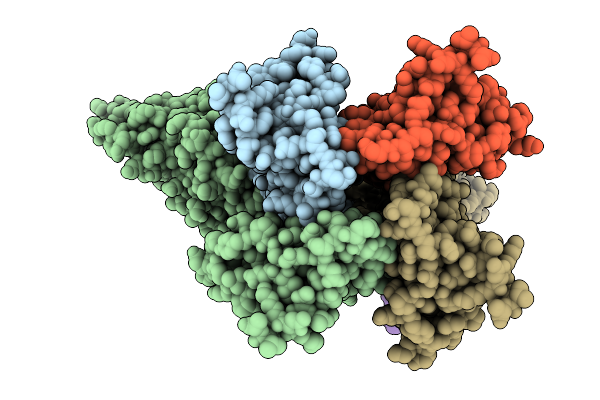
Organism: Salmonella enterica subsp. enterica serovar tennessee, Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: IMMUNE SYSTEM |
 |
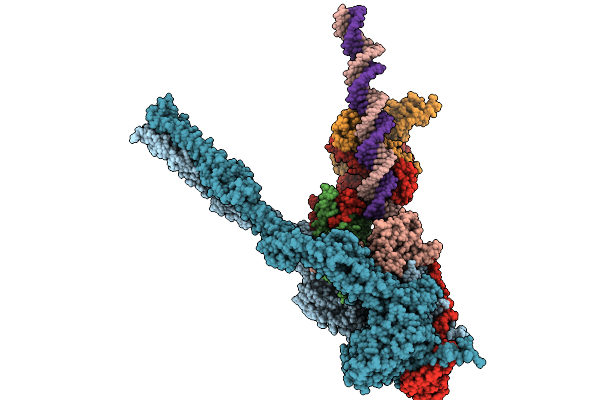
Organism: Salmonella enterica subsp. enterica serovar tennessee, Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: IMMUNE SYSTEM |
 |
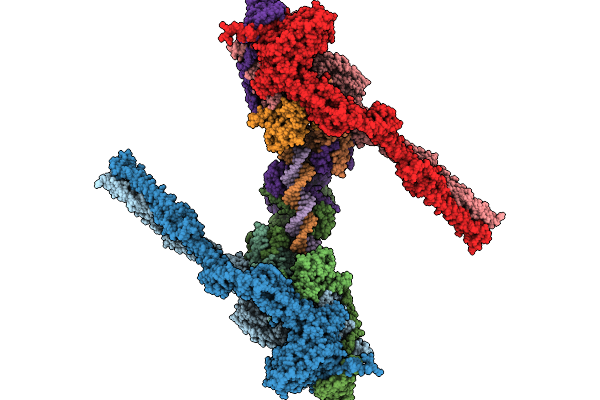
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, BEF |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, BEF |
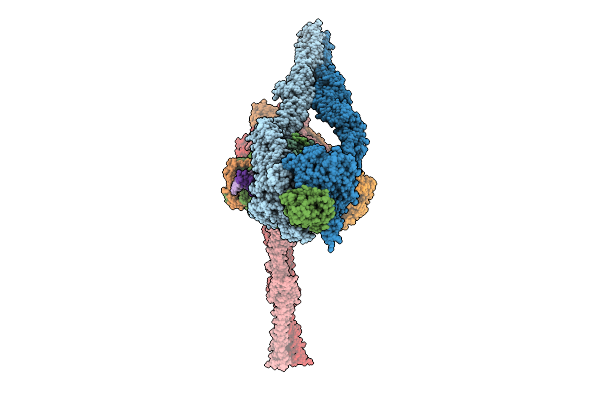 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: DNA BINDING PROTEIN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: DNA BINDING PROTEIN Ligands: ADP |
 |
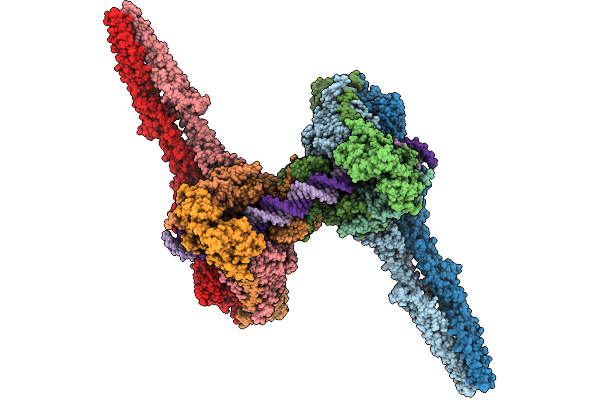
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: DNA BINDING PROTEIN Ligands: ADP |
 |
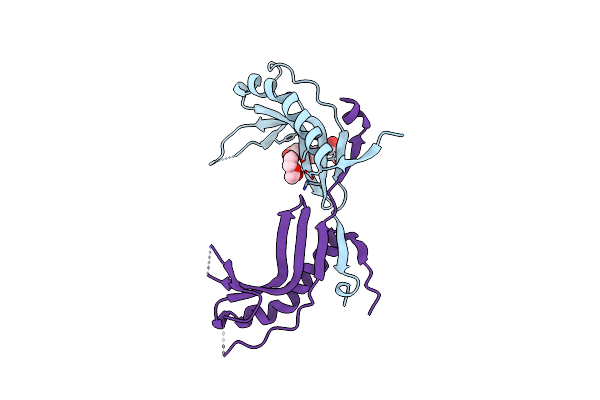
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: DNA BINDING PROTEIN Ligands: ADP |
 |
Crystal Structure In Space Group C2221 Of A Nucleoid-Associated Protein (Ubp) From Sulfolobus Islandicus.
Organism: Sulfolobus islandicus rey15a
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: PEG, PG4 |
 |
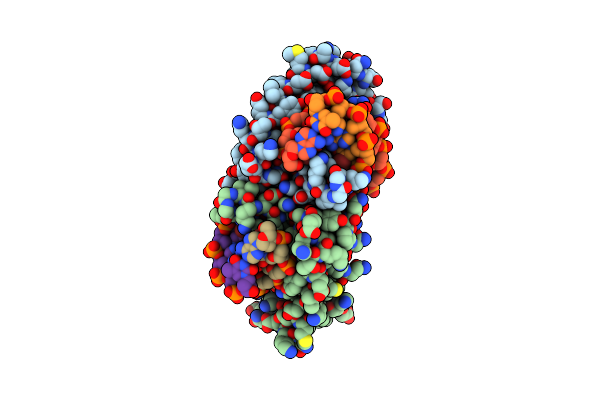
Crystal Structure In Space Group P21 Of A Nucleoid-Associated Protein (Ubp) From Sulfolobus Islandicus.
Organism: Sulfolobus islandicus rey15a
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: PEG, EDO |
 |
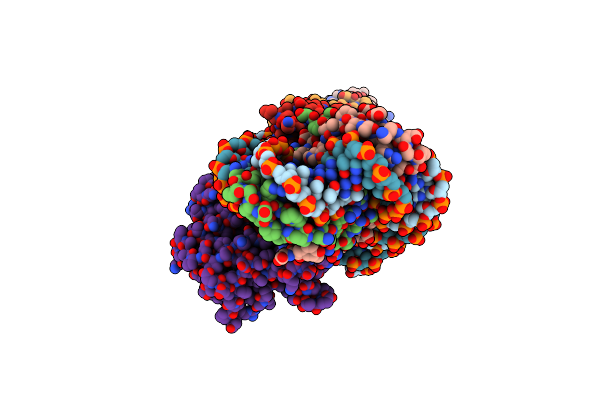
Crystal Structure Of A Nucleoid-Associated Protein (Ubp) Bound To Dna From Sulfolobus Islandicus.
Organism: Sulfolobus islandicus rey15a
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN |
 |
Cryoem Structure Of The F Plasmid Relaxosome In Its Pre-Initiation State, Derived From The Ds-27_+143-R Locally-Refined Map 3.76 A
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN |
 |
Cryoem Structure Of The F Plasmid Relaxosome With Trai In Its Te Mode, Derived From The Ss-27_+8Ds+9_+143-R Locally-Refined 3.45 A Map.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |

