Search Count: 14
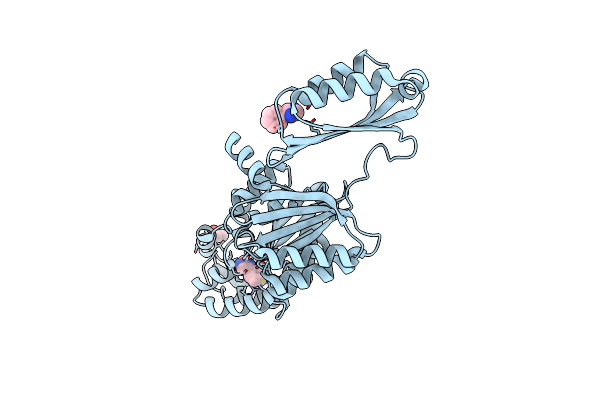 |
Crystal Structure Of Phosphoserine Phosphatase Serb From Mycobacterium Avium In Complex With Phenylimidazole
Organism: Mycobacterium avium 104
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2022-11-23 Classification: HYDROLASE Ligands: MG, PIM, CL, GOL |
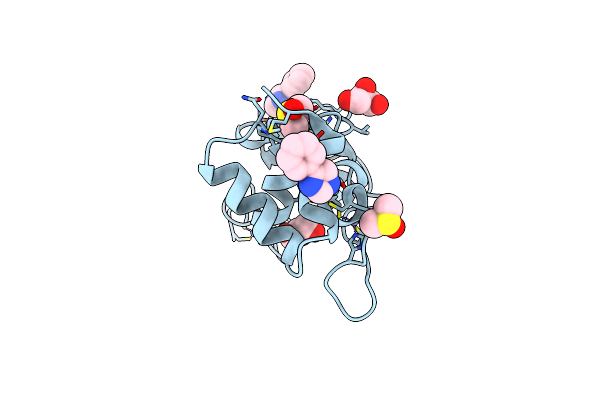 |
Non-Structural Protein 10 (Nsp10) From Sars Cov-2 In Complex With Fragment Vt00022
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2021-10-20 Classification: VIRAL PROTEIN Ligands: ZN, DMS, PIM, GOL, CL |
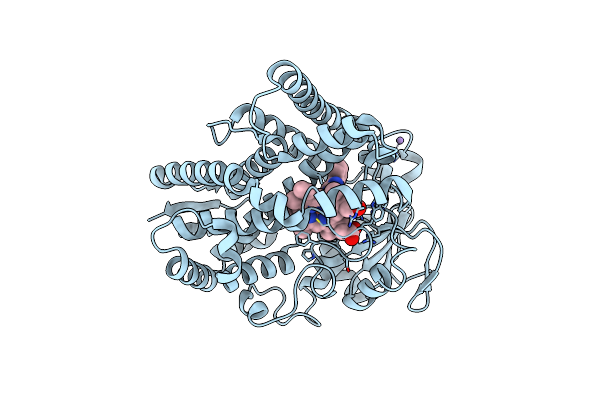 |
Organism: Salvia miltiorrhiza
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-10-24 Classification: OXIDOREDUCTASE Ligands: HEM, PIM, MN |
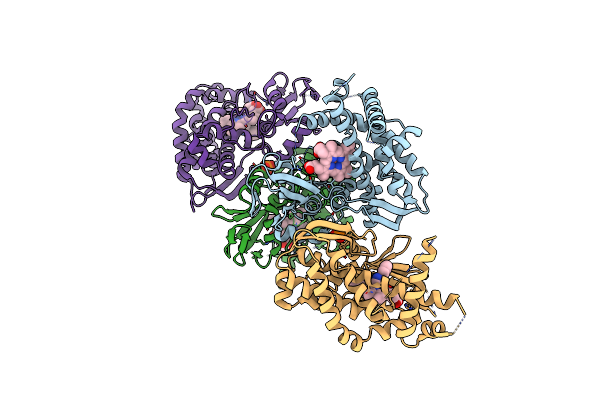 |
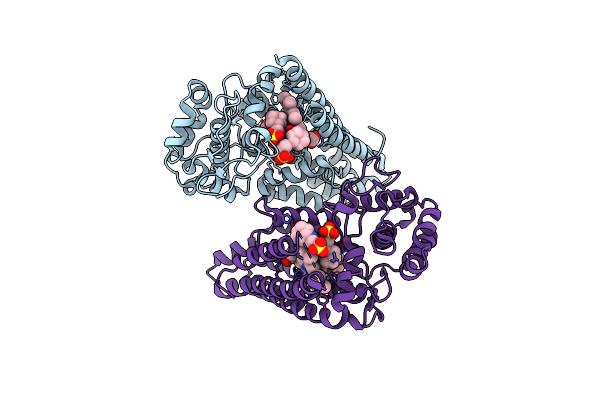
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-02-03 Classification: OXIDOREDUCTASE Ligands: PO4, HEM, PIM |
 |
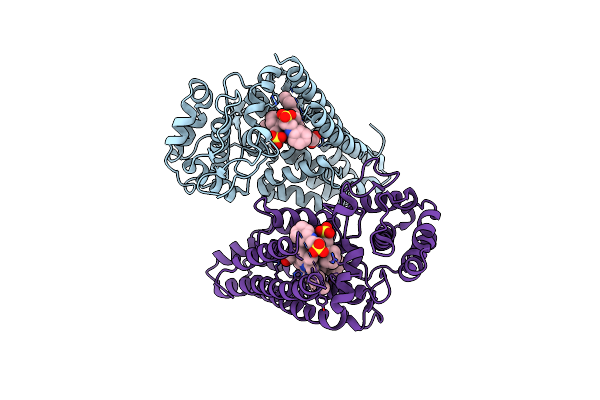
Crystal Structure Of 4-Phenylimidazole Bound Form Of Human Indoleamine 2,3-Dioxygenase (A260G Mutant)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-09-02 Classification: OXIDOREDUCTASE Ligands: HEM, PIM, NHE |
 |
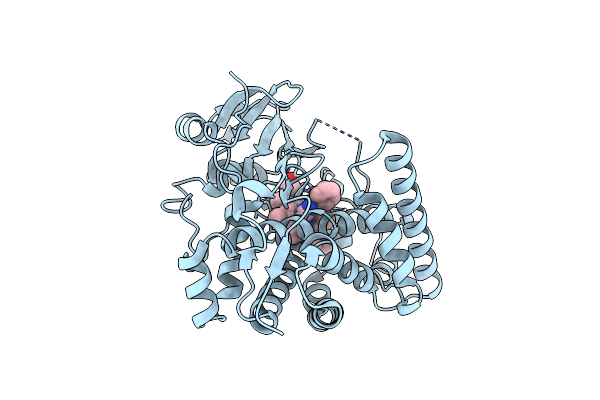
Crystal Structure Of 4-Phenylimidazole Bound Form Of Human Indoleamine 2,3-Dioxygenase (G262A Mutant)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2015-09-02 Classification: OXIDOREDUCTASE Ligands: HEM, PIM, NHE |
 |
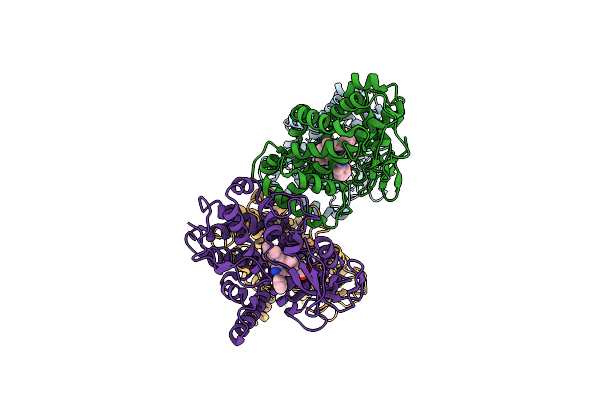
Organism: Streptomyces avermitilis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-12-30 Classification: OXIDOREDUCTASE Ligands: HEM, PIM |
 |
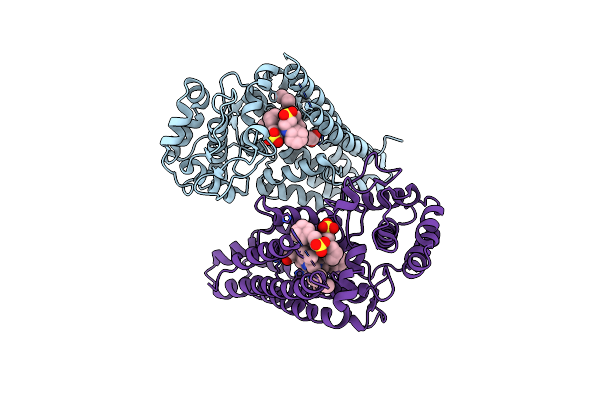
Ligand Bound (4-Phenylimidazole) Crystal Structure Of A Cytochrome P450 From The Thermoacidophilic Archaeon Picrophilus Torridus
Organism: Picrophilus torridus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2008-01-29 Classification: OXIDOREDUCTASE Ligands: HEM, PIM |
 |
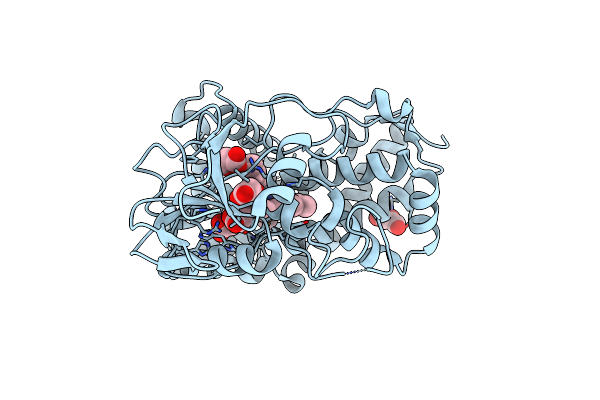
Crystal Structure Of 4-Phenylimidazole Bound Form Of Human Indoleamine 2,3-Dioxygenase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-01-31 Classification: OXIDOREDUCTASE Ligands: HEM, PIM, NHE |
 |
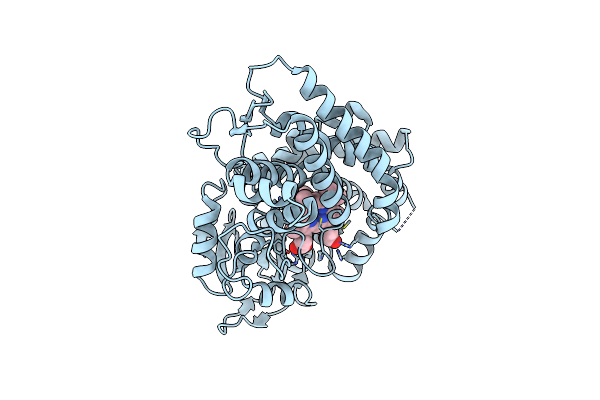
Crystal Structure Of Streptomyces Coelicolor A3(2) Cyp158A2 From Antibiotic Biosynthetic Pathways
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2005-01-11 Classification: OXIDOREDUCTASE Ligands: HG, HEM, PIM, MLA, GOL |
 |
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2004-01-02 Classification: OXIDOREDUCTASE Ligands: HEM, PIM |
 |
Cytochrome P450 14 Alpha-Sterol Demethylase (Cyp51) From Mycobacterium Tuberculosis In Complex With 4-Phenylimidazole
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2000-11-01 Classification: OXIDOREDUCTASE Ligands: HEM, PIM |
 |
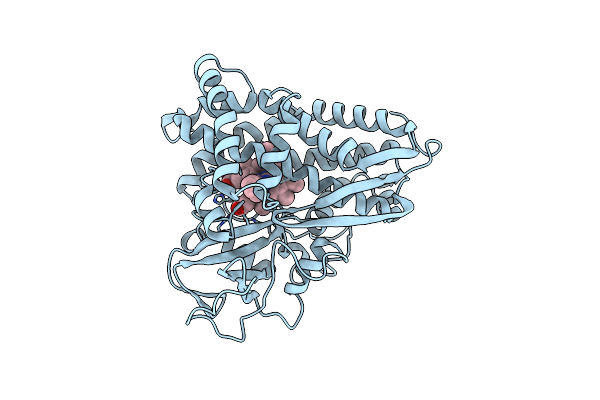
Thermophilic P450: Cyp119 From Sulfolobus Solfactaricus With 4-Phenylimidazole Bound
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2000-10-23 Classification: OXIDOREDUCTASE Ligands: ZN, SO4, HEM, PIM |
 |
Crystal Structures Of Metyrapone-And Phenylimidazole-Inhibited Complexes Of Cytochrome P450-Cam
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 1993-10-31 Classification: OXIDOREDUCTASE(OXYGENASE) Ligands: HEM, PIM |

