Search Count: 6,503
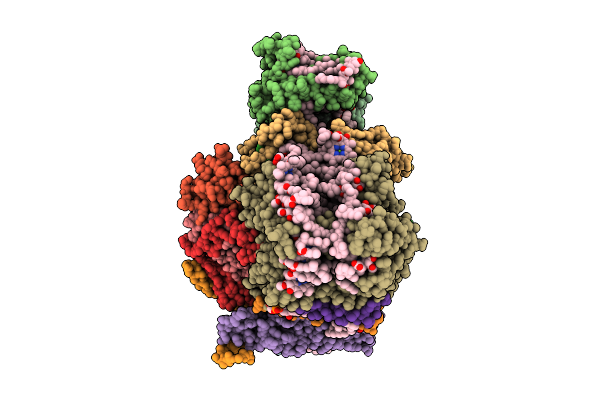 |
Structure Of The Wild-Type Psi-5Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, LMG, A1L1F, PQN, LHG, BCR, SF4 |
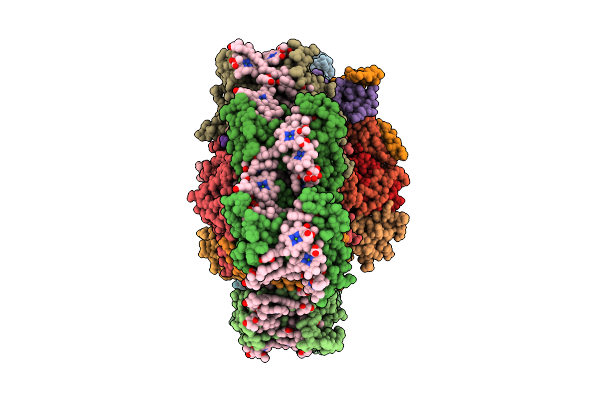 |
Structure Of The Canthaxanthin Mutant Psi-9Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, A1L1F, 45D, LHG, DGD, LMG, PQN, BCR, SF4 |
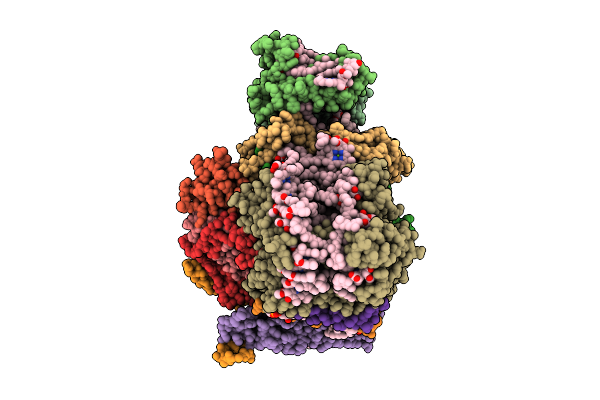 |
Structure Of The Canthaxanthin Mutant Psi-5Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, LMG, A1L1F, PQN, LHG, BCR, SF4 |
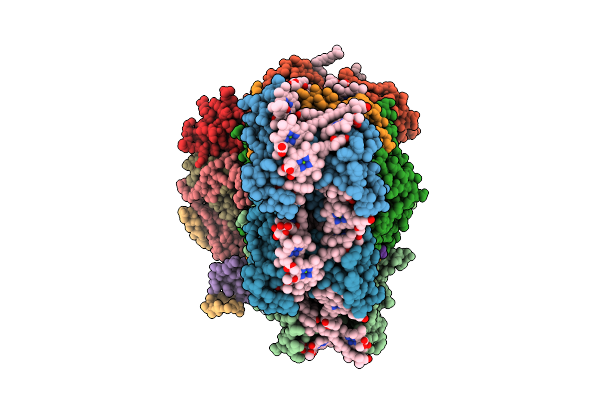 |
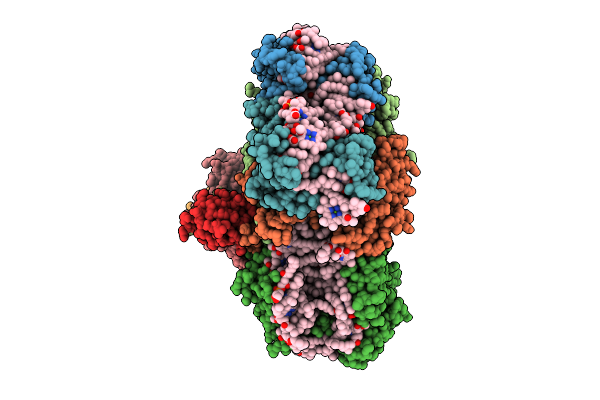
Structure Of The Canthaxanthin Mutant Psi-4Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: A1L1G, A1L1F, XAT, 45D, LHG, CLA, DGD, SQD, PQN, BCR, SF4, LMG |
 |
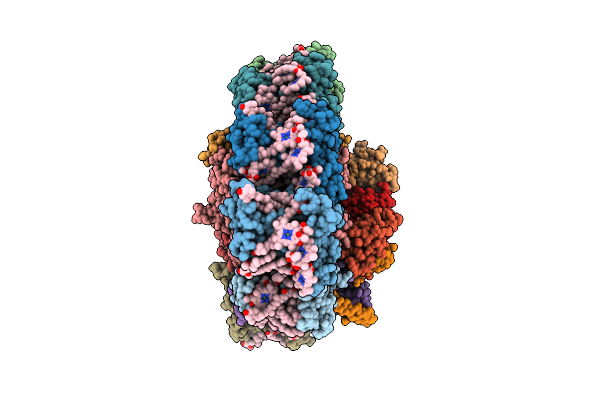
Structure Of The Canthaxanthin Mutant Psi-3Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, A1L1F, PQN, LHG, BCR, SF4, LMG |
 |
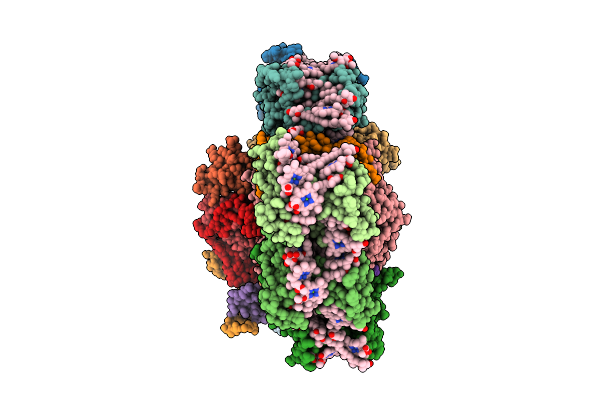
Structure Of The Astaxanthin Mutant Psi-9Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, A1L1F, LHG, DGD, LMG, PQN, BCR, SF4 |
 |
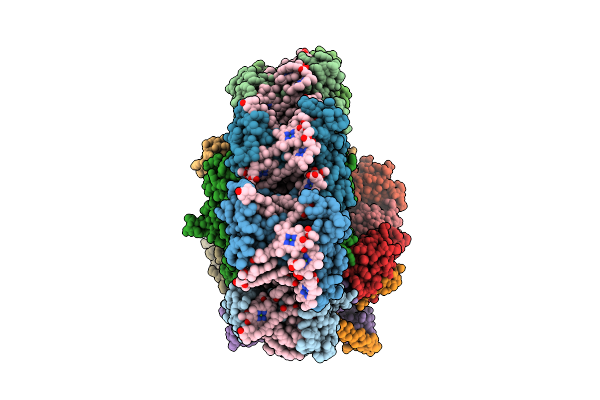
Structure Of The Astaxanthin Mutant Psi-7Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, A1L1F, LHG, DGD, PQN, BCR, SF4, LMG |
 |
Structure Of The Astaxanthin Mutant Psi-5Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, LMG, A1L1F, PQN, LHG, BCR, SF4 |
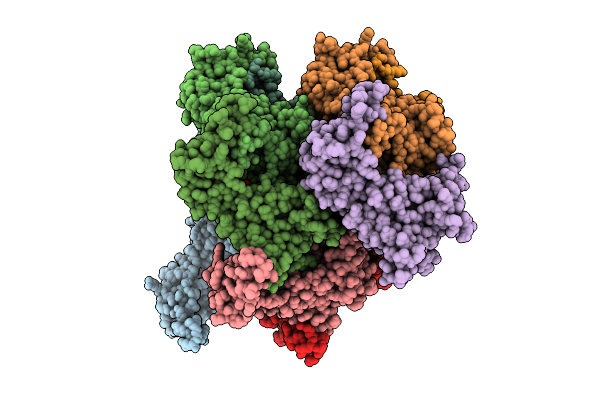 |
Eleven Polymer Msp1 From S.Cerevisiae (With A Catalytic Dead Mutaion) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
Organism: Erythrobacter sanguineus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: BCL, A1ELD |
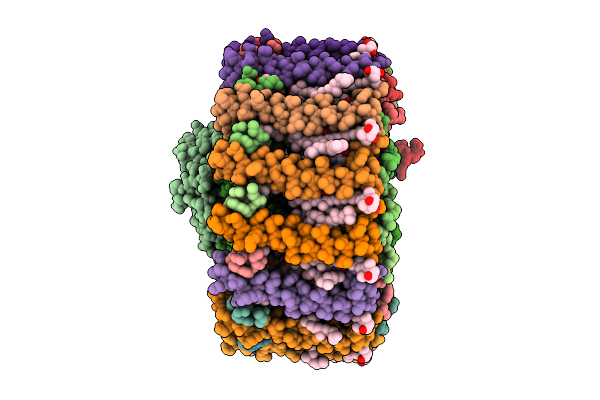 |
Organism: Erythrobacter sanguineus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: BCL, A1EL2, A1ELD, 8K6, PGV, LMT, BPH, UQ8, FE, H4X, PEF, PO4, LHG |
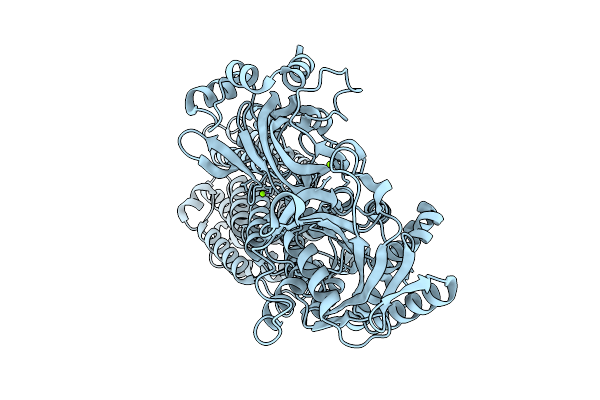 |
Organism: Lactococcus lactis subsp. lactis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: MG |
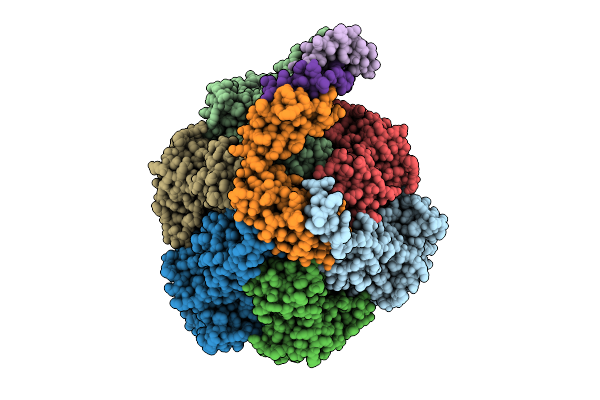 |
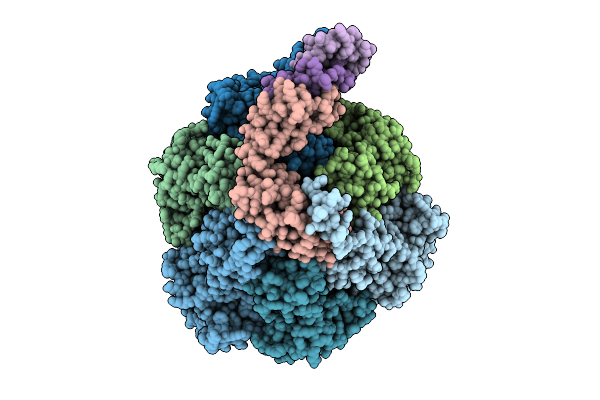
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP, PO4 |
 |
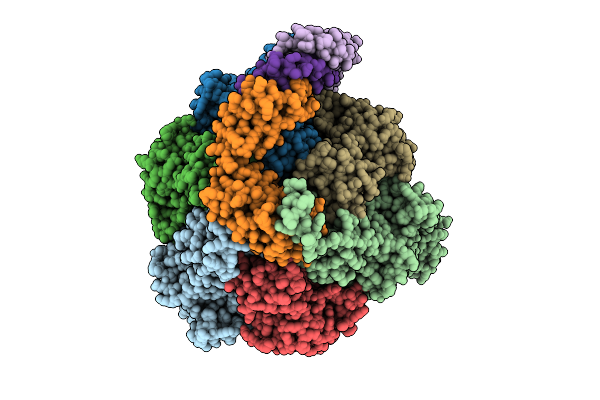
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP, ZN |
 |
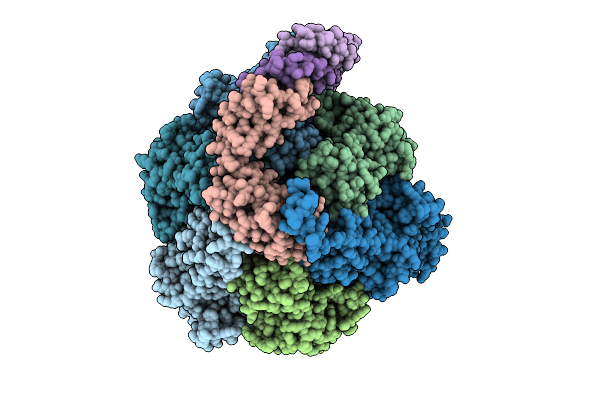
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |
 |
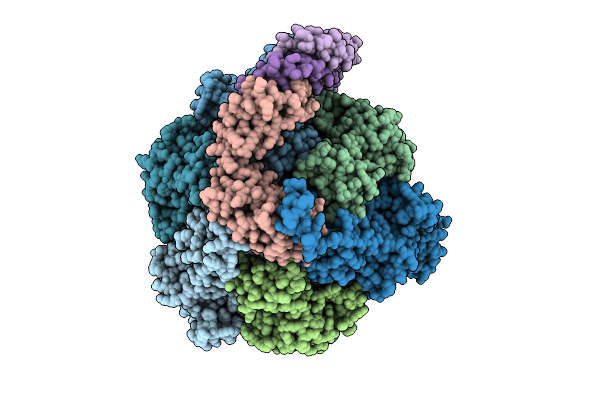
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP, ZN |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |
 |
Structure Of The Wild-Type Psi-9Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, A1L1F, LHG, DGD, LMG, PQN, BCR, SF4 |

