Search Count: 1,847
All
Selected
 |
Organism: Homo sapiens, Bos taurus
Method: ELECTRON MICROSCOPY Resolution:4.40 Å Release Date: 2026-02-18 Classification: SIGNALING PROTEIN Ligands: ME7, CA |
 |
Organism: Homo sapiens, Bos taurus
Method: ELECTRON MICROSCOPY Resolution:7.00 Å Release Date: 2026-02-18 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens, Bos taurus
Method: ELECTRON MICROSCOPY Resolution:4.50 Å Release Date: 2026-02-18 Classification: SIGNALING PROTEIN |
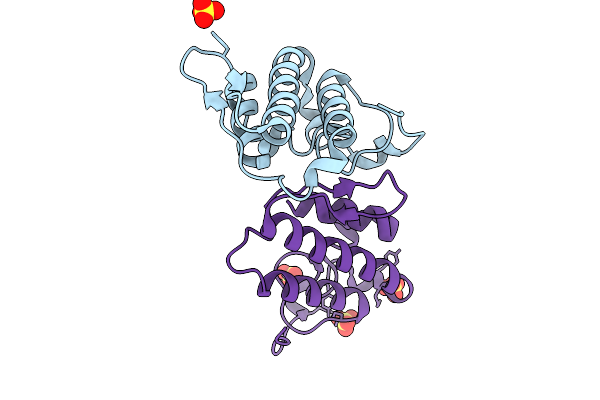 |
Crystal Structure Of Heterodimeric Crotoxin B From Crotalus Durissus Collilineatus
Organism: Crotalus durissus collilineatus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2026-01-28 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Homo sapiens, Micrurus tener tener
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Micrurus tener tener
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN |
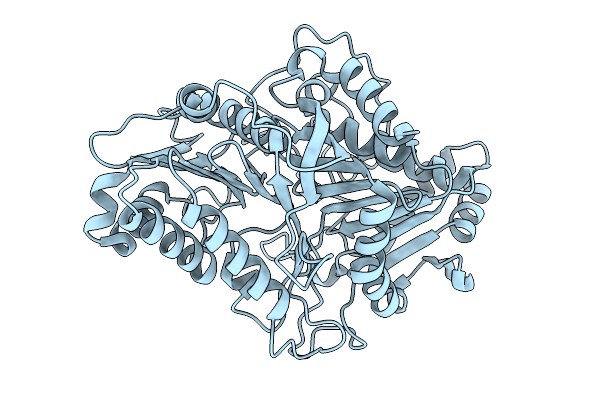 |
Organism: Streptomyces klenkii
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2026-01-21 Classification: HYDROLASE |
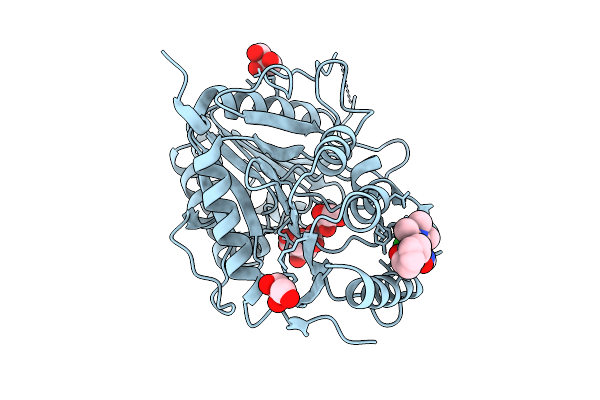 |
Organism: Vaccinia virus western reserve
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: GOL, CIT, A1I9D |
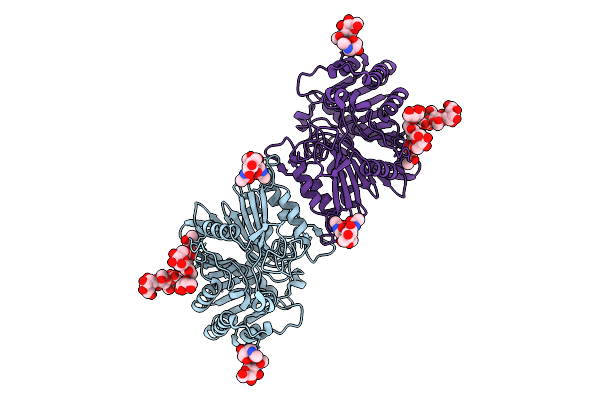 |
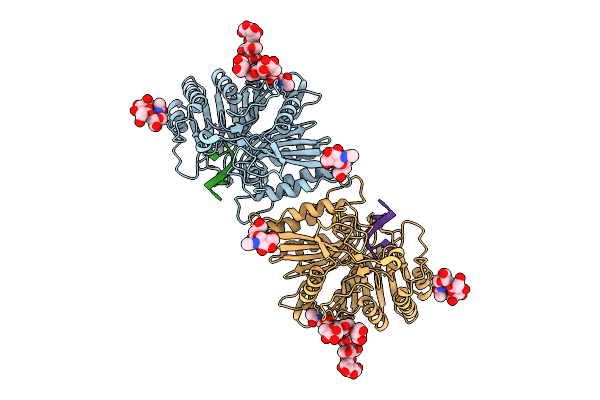
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
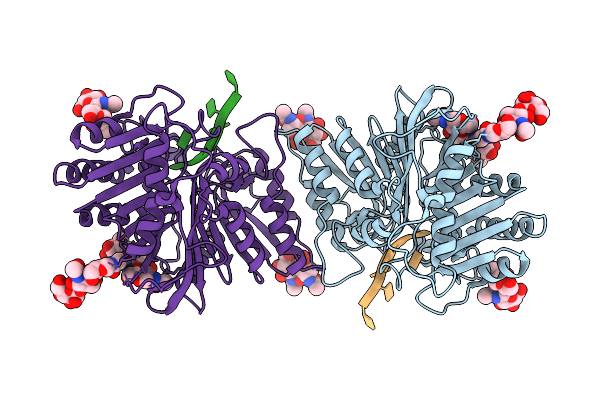
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: IMMUNE SYSTEM |
 |
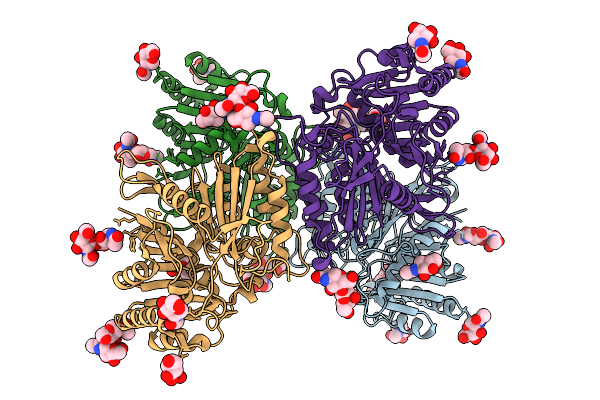
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: IMMUNE SYSTEM |
 |
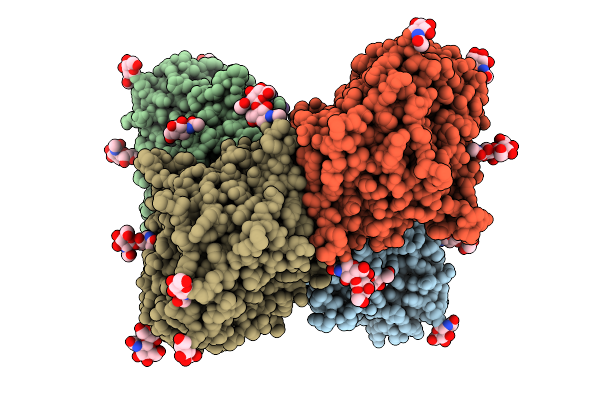
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: IMMUNE SYSTEM Ligands: NAG, T3P |
 |
Structure Of Phospholipase D Betaib1I From Sicarius Terrosus Venom, H47N Mutant Bound To Product And Substrate Sphingolipids At 2.2 A Resolution From A 2-Day Old Crystal
Organism: Sicarius terrosus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-12-03 Classification: LYASE Ligands: A1A43, A1A44, NA, MG, MPD |
 |
Structure Of Phospholipase D Betaib1I From Sicarius Terrosus Venom, H47N Mutant Bound To Substrate Sphingolipids At 2.60 A Resolution
Organism: Sicarius terrosus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-12-03 Classification: LYASE Ligands: A1A43, NA, MG, MPD |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE Ligands: 4BW |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2025-11-19 Classification: HYDROLASE |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-11-19 Classification: HYDROLASE |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Resolution:3.04 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: 4BW |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2025-11-12 Classification: ONCOPROTEIN Ligands: MG, GDP |

